Phosphoregulation of Phase Separation by the SARS-CoV-2 N Protein Suggests a Biophysical Basis for its Dual Functions
- PMID: 33248025
- PMCID: PMC7677695
- DOI: 10.1016/j.molcel.2020.11.025
Phosphoregulation of Phase Separation by the SARS-CoV-2 N Protein Suggests a Biophysical Basis for its Dual Functions
Abstract
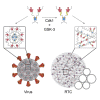
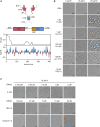
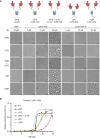
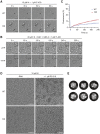
The nucleocapsid (N) protein of coronaviruses serves two major functions: compaction of the RNA genome in the virion and regulation of viral gene transcription. It is not clear how the N protein mediates such distinct functions. The N protein contains two RNA-binding domains surrounded by regions of intrinsic disorder. Phosphorylation of the central disordered region promotes the protein's transcriptional function, but the underlying mechanism is not known. Here, we show that the N protein of SARS-CoV-2, together with viral RNA, forms biomolecular condensates. Unmodified N protein forms partially ordered gel-like condensates and discrete 15-nm particles based on multivalent RNA-protein and protein-protein interactions. Phosphorylation reduces these interactions, generating a more liquid-like droplet. We propose that distinct oligomeric states support the two functions of the N protein: unmodified protein forms a structured oligomer that is suited for nucleocapsid assembly, and phosphorylated protein forms a liquid-like compartment for viral genome processing.
Keywords: COVID-19; Coronavirus; N protein; SARS-CoV-2; biomolecular condensate; nucleocapsid; phase separation; phosphorylation.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures

Update of
-
Phosphorylation modulates liquid-liquid phase separation of the SARS-CoV-2 N protein.bioRxiv [Preprint]. 2020 Jun 29:2020.06.28.176248. doi: 10.1101/2020.06.28.176248. bioRxiv. 2020. Update in: Mol Cell. 2020 Dec 17;80(6):1092-1103.e4. doi: 10.1016/j.molcel.2020.11.025. PMID: 32637943 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

