Multi-omics integration in biomedical research - A metabolomics-centric review
- PMID: 33248648
- PMCID: PMC7701361
- DOI: 10.1016/j.aca.2020.10.038
Multi-omics integration in biomedical research - A metabolomics-centric review
Abstract
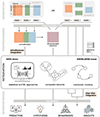
Recent advances in high-throughput technologies have enabled the profiling of multiple layers of a biological system, including DNA sequence data (genomics), RNA expression levels (transcriptomics), and metabolite levels (metabolomics). This has led to the generation of vast amounts of biological data that can be integrated in so-called multi-omics studies to examine the complex molecular underpinnings of health and disease. Integrative analysis of such datasets is not straightforward and is particularly complicated by the high dimensionality and heterogeneity of the data and by the lack of universal analysis protocols. Previous reviews have discussed various strategies to address the challenges of data integration, elaborating on specific aspects, such as network inference or feature selection techniques. Thereby, the main focus has been on the integration of two omics layers in their relation to a phenotype of interest. In this review we provide an overview over a typical multi-omics workflow, focusing on integration methods that have the potential to combine metabolomics data with two or more omics. We discuss multiple integration concepts including data-driven, knowledge-based, simultaneous and step-wise approaches. We highlight the application of these methods in recent multi-omics studies, including large-scale integration efforts aiming at a global depiction of the complex relationships within and between different biological layers without focusing on a particular phenotype.
Keywords: Data integration; Lipidomics; Metabolomics; Multi-omics; Systems biology.
Copyright © 2020 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

References
-
- Lotta LA, Scott RA, Sharp SJ, Burgess S, Luan J, Tillin T, Schmidt AF, Imamura F, Stewart ID, Perry JRB, Marney L, Koulman A, Karoly ED, Forouhi NG, Sjögren RJO, Näslund E, Zierath JR, Krook A, Savage DB, Griffin JL, Chaturvedi N, Hingorani AD, Khaw KT, Barroso I, McCarthy MI, O’Rahilly S, Wareham NJ, Langenberg C, Genetic Predisposition to an Impaired Metabolism of the Branched-Chain Amino Acids and Risk of Type 2 Diabetes: A Mendelian Randomisation Analysis, PLoS Med. 13 (2016) 1–22. 10.1371/journal.pmed.1002179. - DOI - PMC - PubMed
-
- Tynkkynen J, Chouraki V, van der Lee SJ, Hernesniemi J, Yang Q, Li S, Beiser A, Larson MG, Sääksjärvi K, Shipley MJ, Singh-Manoux A, Gerszten RE, Wang TJ, Havulinna AS, Würtz P, Fischer K, Demirkan A, Ikram MA, Amin N, Lehtimäki T, Kähönen M, Perola M, Metspalu A, Kangas AJ, Soininen P, Ala-Korpela M, Vasan RS, Kivimäki M, van Duijn CM, Seshadri S, Salomaa V, Association of branched-chain amino acids and other circulating metabolites with risk of incident dementia and Alzheimer’s disease: A prospective study in eight cohorts, Alzheimer’s Dement. 14 (2018) 723–733. 10.1016/j.jalz.2018.01.003. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

