Recurrent Topics in Mass Spectrometry-Based Metabolomics and Lipidomics-Standardization, Coverage, and Throughput
- PMID: 33249827
- PMCID: PMC7807424
- DOI: 10.1021/acs.analchem.0c04698
Recurrent Topics in Mass Spectrometry-Based Metabolomics and Lipidomics-Standardization, Coverage, and Throughput
Conflict of interest statement
The authors declare no competing financial interest.
Figures


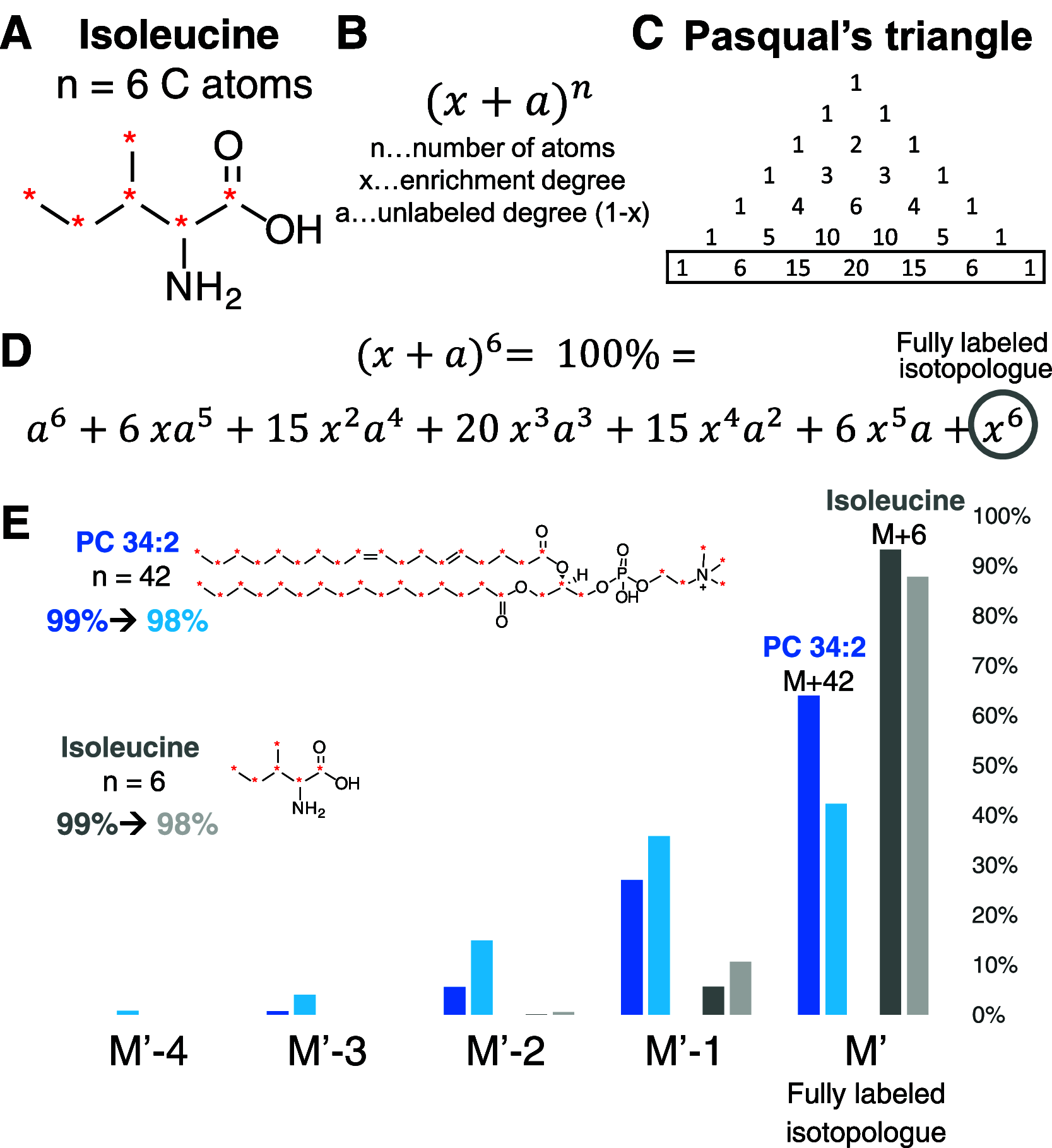



References
-
- González-Riano C.; Dudzik D.; Garcia A.; Gil-de-la-Fuente A.; Gradillas A.; Godzien J.; López-Gonzálvez Á.; Rey-Stolle F.; Rojo D.; Ruperez F. J.; Saiz J.; Barbas C. Recent Developments along the Analytical Process for Metabolomics Workflows. Anal. Chem. 2020, 92 (1), 203–226. 10.1021/acs.analchem.9b04553. - DOI - PubMed
-
- Dunn W. B.; Ellis D. I. Metabolomics: Current Analytical Platforms and Methodologies. TrAC, Trends Anal. Chem. 2005, 24 (4), 285–294. 10.1016/j.trac.2004.11.021. - DOI
-
- Lerma-Ortiz C.; Jeffryes J. G.; Cooper A. J. L.; Niehaus T. D.; Thamm A. M. K.; Frelin O.; Aunins T.; Fiehn O.; de Crécy-Lagard V.; Henry C. S.; Hanson A. D. ‘Nothing of Chemistry Disappears in Biology’: The Top 30 Damage-Prone Endogenous Metabolites. Biochem. Soc. Trans. 2016, 44 (3), 961–971. 10.1042/BST20160073. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

