Proficiency Testing of Metagenomics-Based Detection of Food-Borne Pathogens Using a Complex Artificial Sequencing Dataset
- PMID: 33250869
- PMCID: PMC7672002
- DOI: 10.3389/fmicb.2020.575377
Proficiency Testing of Metagenomics-Based Detection of Food-Borne Pathogens Using a Complex Artificial Sequencing Dataset
Abstract
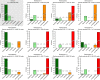
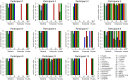
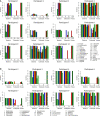
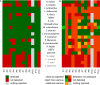
Metagenomics-based high-throughput sequencing (HTS) enables comprehensive detection of all species comprised in a sample with a single assay and is becoming a standard method for outbreak investigation. However, unlike real-time PCR or serological assays, HTS datasets generated for pathogen detection do not easily provide yes/no answers. Rather, results of the taxonomic read assignment need to be assessed by trained personnel to gain information thereof. Proficiency tests are important instruments of validation, harmonization, and standardization. Within the European Union funded project COMPARE [COllaborative Management Platform for detection and Analyses of (Re-) emerging and foodborne outbreaks in Europe], we conducted a proficiency test to scrutinize the ability to assess diagnostic metagenomics data. An artificial dataset resembling shotgun sequencing of RNA from a sample of contaminated trout was provided to 12 participants with the request to provide a table with per-read taxonomic assignments at species level and a report with a summary and assessment of their findings, considering different categories like pathogen, background, or contaminations. Analysis of the read assignment tables showed that the software used reliably classified the reads taxonomically overall. However, usage of incomplete reference databases or inappropriate data pre-processing caused difficulties. From the combination of the participants' reports with their read assignments, we conclude that, although most species were detected, a number of important taxa were not or not correctly categorized. This implies that knowledge of and awareness for potentially dangerous species and contaminations need to be improved, hence, capacity building for the interpretation of diagnostic metagenomics datasets is necessary.
Keywords: background contamination; diagnostic assessment; high-throughput sequencing; metagenomics; pathogen; proficiency test; training.
Copyright © 2020 Höper, Grützke, Brinkmann, Mossong, Matamoros, Ellis, Deneke, Tausch, Cuesta, Monzón, Juliá, Petersen, Hendriksen, Pamp, Leijon, Hakhverdyan, Walsh, Cotter, Chandrasekaran, Tay, Schlundt, Sala, De Cesare, Nitsche, Beer and Wylezich.
Figures
References
LinkOut - more resources
Full Text Sources

