Phylogenomics of the Andean Tetraploid Clade of the American Amaryllidaceae (Subfamily Amaryllidoideae): Unlocking a Polyploid Generic Radiation Abetted by Continental Geodynamics
- PMID: 33250911
- PMCID: PMC7674842
- DOI: 10.3389/fpls.2020.582422
Phylogenomics of the Andean Tetraploid Clade of the American Amaryllidaceae (Subfamily Amaryllidoideae): Unlocking a Polyploid Generic Radiation Abetted by Continental Geodynamics
Erratum in
-
Corrigendum: Phylogenomics of the Andean tetraploid clade of the American Amaryllidaceae (subfamily Amaryllidoideae): Unlocking a polyploid generic radiation abetted by continental geodynamics.Front Plant Sci. 2023 Mar 2;14:1151864. doi: 10.3389/fpls.2023.1151864. eCollection 2023. Front Plant Sci. 2023. PMID: 36938047 Free PMC article.
Abstract
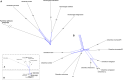
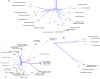
One of the two major clades of the endemic American Amaryllidaceae subfam. Amaryllidoideae constitutes the tetraploid-derived (n = 23) Andean-centered tribes, most of which have 46 chromosomes. Despite progress in resolving phylogenetic relationships of the group with plastid and nrDNA, certain subclades were poorly resolved or weakly supported in those previous studies. Sequence capture using anchored hybrid enrichment was employed across 95 species of the clade along with five outgroups and generated sequences of 524 nuclear genes and a partial plastome. Maximum likelihood phylogenetic analyses were conducted on concatenated supermatrices, and coalescent-based species tree analyses were run on the gene trees, followed by hybridization network, age diversification and biogeographic analyses. The four tribes Clinantheae, Eucharideae, Eustephieae, and Hymenocallideae (sister to Clinantheae) are resolved in all analyses with > 90 and mostly 100% support, as are almost all genera within them. Nuclear gene supermatrix and species tree results were largely in concordance; however, some instances of cytonuclear discordance were evident. Hybridization network analysis identified significant reticulation in Clinanthus, Hymenocallis, Stenomesson and the subclade of Eucharideae comprising Eucharis, Caliphruria, and Urceolina. Our data support a previous treatment of the latter as a single genus, Urceolina, with the addition of Eucrosia dodsonii. Biogeographic analysis and penalized likelihood age estimation suggests an origin in the Cauca, Desert and Puna Neotropical bioprovinces for the complex in the mid-Oligocene, with more dispersals than vicariances in its history, but no extinctions. Hymenocallis represents the only instance of long-distance vicariance from the tropical Andean origin of its tribe Hymenocallideae. The absence of extinctions correlates with the lack of diversification rate shifts within the clade. The Eucharideae experienced a sudden lineage radiation ca. 10 Mya. We tie much of the divergences in the Andean-centered lineages to the rise of the Andes, and suggest that the Amotape-Huancabamba Zone functioned as both a corridor (dispersal) and a barrier to migration (vicariance). Several taxonomic changes are made. This is the largest DNA sequence data set to be applied within Amaryllidaceae to date.
Keywords: Andes; Asparagales; anchored hybrid enrichment; biogeography; geophyte; molecular systematics; monocotyledons; phylogenetics.
Copyright © 2020 Meerow, Gardner and Nakamura.
Figures








References
-
- Anthelme F., Jacobsen D., Macek P., Meneses R. I., Moret P., Beck S., et al. (2014). Biodiversity patterns and continental insularity in the tropical high Andes. AAAR 46 811–828. 10.1657/1938-4246-46.4.811 - DOI
Associated data
LinkOut - more resources
Full Text Sources
Miscellaneous

