Pnictogen-bonding catalysis: brevetoxin-type polyether cyclizations
- PMID: 33250977
- PMCID: PMC7690316
- DOI: 10.1039/d0sc02551h
Pnictogen-bonding catalysis: brevetoxin-type polyether cyclizations
Abstract
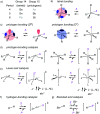
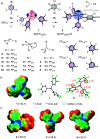
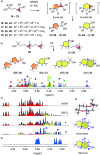
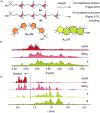
Pnictogen-bond donors are attractive for use in catalysis because of deep σ holes, high multivalency, rich hypervalency, and chiral binding pockets. We here report natural product inspired epoxide-opening polyether cyclizations catalyzed by fluoroarylated Sb(v) > Sb(iii) > Bi > Sn > Ge. The distinctive characteristic found for pnictogen-bonding catalysis is the breaking of the Baldwin rules, that is selective endo cyclization into the trans-fused ladder oligomers known from the brevetoxins. Moreover, tris(3,4,5-trifluorophenyl)stibines and their hypervalent stiborane catecholates afford different anti-Baldwin stereoselectivity. Lewis (SbCl3), Brønsted (AcOH) and π acids fail to provide similar access to these forbidden rings. Like hydrogen-bonding catalysis differs from Brønsted acid catalysis, pnictogen-bonding catalysis thus emerges as the supramolecular counterpart of covalent Lewis acid catalysis.
This journal is © The Royal Society of Chemistry 2020.
Figures
References
-
- Alkorta I., Elguero J., Frontera A. Crystals. 2020;10:180.
- Bamberger J., Ostler F., Garcia Mancheño O. ChemCatChem. 2019;11:5198–5211. - PMC - PubMed
- Sutar R. L., Huber M. S. ACS Catal. 2019;9:9622–9639.
- Taylor M. S. Coord. Chem. Rev. 2020;413:213270.
- Lim J. Y. C., Beer P. D. Chem. 2018;4:731–783.
- Scheiner S., Lu J. Chem.–Eur. J. 2018;24:8167–8177. - PubMed
-
- Scilabra P., Terraneo G., Resnati G. J. Fluorine Chem. 2017;203:62–74.
- Scheiner S. Acc. Chem. Res. 2013;46:280–288. - PubMed
- Moaven S., Yu J., Vega M., Unruh D. K., Cozzolino A. F. Chem. Commun. 2018;54:8849–8852. - PubMed
- Leroy C., Johannson R., Bryce D. L. J. Phys. Chem. A. 2019;123:1030–1043. - PubMed
- Bauzá A., Mooibroek T. J., Frontera A. ChemPhysChem. 2016;17:1608–1614. - PubMed
- Girolami G. S. J. Chem. Educ. 2009;86:1200.
- Starbuck J., Norman N. C., Orpen A. G. New J. Chem. 1999;23:969–972.
-
- Bauzá A., Mooibroek T. J., Frontera A. Angew. Chem., Int. Ed. 2013;52:12317–12321. - PubMed
- Karim A., Schulz N., Andersson H., Nekoueishahraki B., Carlsson A.-C. C., Sarabi D., Valkonen A., Rissanen K., Gräfenstein J., Keller S., Erdelyi M. J. Am. Chem. Soc. 2018;140:17571–17579. - PubMed
- Grabowski S. J. Phys. Chem. Chem. Phys. 2014;16:1824–1834. - PubMed
- Michalczyk M., Zierkiewicz W., Wysokiński R., Scheiner S. ChemPhysChem. 2019;20:959–966. - PubMed
- Scheiner S. J. Phys. Chem. A. 2018;122:2550–2562. - PubMed
- Nakamura H., Ishihara K., Yamamoto H. J. Org. Chem. 2002;67:5124–5137. - PubMed
- Berger R., Duff K., Leighton J. L. J. Am. Chem. Soc. 2004;126:5686–5687. - PubMed
- Hrdina R., Müller C. E., Wende R. C., Lippert K. M., Benassi M., Spengler B., Schreiner P. R. J. Am. Chem. Soc. 2011;133:7624–7627. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

