Combinations of PCR and Isothermal Amplification Techniques Are Suitable for Fast and Sensitive Detection of SARS-CoV-2 Viral RNA
- PMID: 33251206
- PMCID: PMC7672014
- DOI: 10.3389/fbioe.2020.604793
Combinations of PCR and Isothermal Amplification Techniques Are Suitable for Fast and Sensitive Detection of SARS-CoV-2 Viral RNA
Abstract
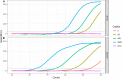
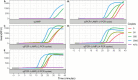
The newly identified coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), causes coronavirus disease 2019 (COVID-19) and has affected over 25 million people worldwide as of August 31, 2020. To aid in the development of diagnostic kits for rapid and sensitive detection of the virus, we evaluated a combination of polymerase chain reaction (PCR) and isothermal nucleic acid amplification techniques. Here, we compared conventional PCR and loop-mediated isothermal amplification (LAMP) methods with hybrid techniques such as polymerase chain displacement reaction (PCDR) and a newly developed PCR-LAMP method. We found that the hybrid methods demonstrated higher sensitivity and assay reaction rates than those of the classic LAMP and PCR techniques and can be used to for SARS-CoV-2 detection. The proposed methods based on the modern hybrid amplification techniques markedly improve virus detection and, therefore, can be extremely useful in the development of new diagnostic kits.
Keywords: COVID-19; PCR-LAMP; SARS-CoV-2; hybrid virus detection technique; loop-mediated isothermal amplification; polymerase chain displacement reaction.
Copyright © 2020 Varlamov, Blagodatskikh, Smirnova, Kramarov and Ignatov.
Figures
Similar articles
-
Loop mediated isothermal amplification (LAMP) assays as a rapid diagnostic for COVID-19.Med Hypotheses. 2020 Aug;141:109786. doi: 10.1016/j.mehy.2020.109786. Epub 2020 Apr 25. Med Hypotheses. 2020. PMID: 32361529 Free PMC article.
-
Development and Clinical Application of a Rapid and Sensitive Loop-Mediated Isothermal Amplification Test for SARS-CoV-2 Infection.mSphere. 2020 Aug 26;5(4):e00808-20. doi: 10.1128/mSphere.00808-20. mSphere. 2020. PMID: 32848011 Free PMC article.
-
A New Method to Detect Variants of SARS-CoV-2 Using Reverse Transcription Loop-Mediated Isothermal Amplification Combined with a Bioluminescent Assay in Real Time (RT-LAMP-BART).Int J Mol Sci. 2023 Jun 27;24(13):10698. doi: 10.3390/ijms241310698. Int J Mol Sci. 2023. PMID: 37445876 Free PMC article.
-
Nucleic Acid Testing of SARS-CoV-2.Int J Mol Sci. 2021 Jun 7;22(11):6150. doi: 10.3390/ijms22116150. Int J Mol Sci. 2021. PMID: 34200331 Free PMC article. Review.
-
A loop-mediated isothermal amplification-enabled analytical assay for the detection of SARS-CoV-2: A review.Front Cell Infect Microbiol. 2022 Dec 23;12:1068015. doi: 10.3389/fcimb.2022.1068015. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36619749 Free PMC article. Review.
Cited by
-
Types and Applications of Nicking Enzyme-Combined Isothermal Amplification.Int J Mol Sci. 2022 Apr 21;23(9):4620. doi: 10.3390/ijms23094620. Int J Mol Sci. 2022. PMID: 35563012 Free PMC article. Review.
-
Rapid Detection of Attomolar SARS-CoV-2 Nucleic Acids in All-Dielectric Metasurface Biosensors.Biosensors (Basel). 2022 Nov 8;12(11):987. doi: 10.3390/bios12110987. Biosensors (Basel). 2022. PMID: 36354496 Free PMC article.
-
Spike protein of SARS-CoV-2 variants: a brief review and practical implications.Braz J Microbiol. 2022 Sep;53(3):1133-1157. doi: 10.1007/s42770-022-00743-z. Epub 2022 Apr 9. Braz J Microbiol. 2022. PMID: 35397075 Free PMC article. Review.
-
BRET-based biosensors for SARS-CoV-2 oligonucleotide detection.Front Bioeng Biotechnol. 2024 Jun 3;12:1353479. doi: 10.3389/fbioe.2024.1353479. eCollection 2024. Front Bioeng Biotechnol. 2024. PMID: 38887615 Free PMC article.
-
COVID-19 Diagnosis: Current and Future Techniques.Int J Biol Macromol. 2021 Dec 15;193(Pt B):1835-1844. doi: 10.1016/j.ijbiomac.2021.11.016. Epub 2021 Nov 12. Int J Biol Macromol. 2021. PMID: 34774862 Free PMC article. Review.
References
-
- Ben-Assa N., Naddaf R., Gefen T., Capucha T., Hajjo H., Mandelbaum N., et al. (2020). SARS-CoV-2 on-the-spot virus detection directly from patients. medRxiv [Preprint]. 10.1101/2020.04.22.20072389 - DOI
-
- Bruce E. A., Tighe S., Hoffman J. J., Laaguiby P., Gerrard D. L., Diehl S. A., et al. (2020). RT-qPCR Detection of SARS-CoV-2 RNA from patient nasopharyngeal swab using qiagen rneasy kits or directly via omission of an rna extraction step. bioRxiv [Preprint]. 10.1101/2020.03.20.001008 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

