Genome-Scale Metabolic Modeling for Unraveling Molecular Mechanisms of High Threat Pathogens
- PMID: 33251208
- PMCID: PMC7673413
- DOI: 10.3389/fcell.2020.566702
Genome-Scale Metabolic Modeling for Unraveling Molecular Mechanisms of High Threat Pathogens
Abstract
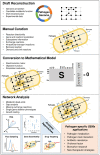
Pathogens give rise to a wide range of diseases threatening global health and hence drawing public health agencies' attention to establish preventative and curative solutions. Genome-scale metabolic modeling is ever increasingly used tool for biomedical applications including the elucidation of antibiotic resistance, virulence, single pathogen mechanisms and pathogen-host interaction systems. With this approach, the sophisticated cellular system of metabolic reactions inside the pathogens as well as between pathogen and host cells are represented in conjunction with their corresponding genes and enzymes. Along with essential metabolic reactions, alternate pathways and fluxes are predicted by performing computational flux analyses for the growth of pathogens in a very short time. The genes or enzymes responsible for the essential metabolic reactions in pathogen growth are regarded as potential drug targets, as a priori guide to researchers in the pharmaceutical field. Pathogens alter the key metabolic processes in infected host, ultimately the objective of these integrative constraint-based context-specific metabolic models is to provide novel insights toward understanding the metabolic basis of the acute and chronic processes of infection, revealing cellular mechanisms of pathogenesis, identifying strain-specific biomarkers and developing new therapeutic approaches including the combination drugs. The reaction rates predicted during different time points of pathogen development enable us to predict active pathways and those that only occur during certain stages of infection, and thus point out the putative drug targets. Among others, fatty acid and lipid syntheses reactions are recent targets of new antimicrobial drugs. Genome-scale metabolic models provide an improved understanding of how intracellular pathogens utilize the existing microenvironment of the host. Here, we reviewed the current knowledge of genome-scale metabolic modeling in pathogen cells as well as pathogen host interaction systems and the promising applications in the extension of curative strategies against pathogens for global preventative healthcare.
Keywords: flux balance analysis (FBA); flux variability analysis; gene essentiality; genome-scale metabolic models; infection; pathogen; pathogen-host interactions; systems biology.
Copyright © 2020 Sertbas and Ulgen.
Figures


Similar articles
-
Editorial: Current status and perspective on drug targets in tubercle bacilli and drug design of antituberculous agents based on structure-activity relationship.Curr Pharm Des. 2014;20(27):4305-6. doi: 10.2174/1381612819666131118203915. Curr Pharm Des. 2014. PMID: 24245755
-
Integration of Metabolomics and Transcriptomics Reveals a Complex Diet of Mycobacterium tuberculosis during Early Macrophage Infection.mSystems. 2017 Aug 22;2(4):e00057-17. doi: 10.1128/mSystems.00057-17. eCollection 2017 Jul-Aug. mSystems. 2017. PMID: 28845460 Free PMC article.
-
Integrated Metabolic Modeling, Culturing, and Transcriptomics Explain Enhanced Virulence of Vibrio cholerae during Coinfection with Enterotoxigenic Escherichia coli.mSystems. 2020 Sep 8;5(5):e00491-20. doi: 10.1128/mSystems.00491-20. mSystems. 2020. PMID: 32900868 Free PMC article.
-
Computational Systems Biology of Metabolism in Infection.Exp Suppl. 2018;109:235-282. doi: 10.1007/978-3-319-74932-7_6. Exp Suppl. 2018. PMID: 30535602 Review.
-
Biomedical applications of genome-scale metabolic network reconstructions of human pathogens.Curr Opin Biotechnol. 2018 Jun;51:70-79. doi: 10.1016/j.copbio.2017.11.014. Epub 2017 Dec 7. Curr Opin Biotechnol. 2018. PMID: 29223465 Free PMC article. Review.
Cited by
-
Mathematical models to study the biology of pathogens and the infectious diseases they cause.iScience. 2022 Mar 15;25(4):104079. doi: 10.1016/j.isci.2022.104079. eCollection 2022 Apr 15. iScience. 2022. PMID: 35359802 Free PMC article. Review.
-
Systems Biology: New Insight into Antibiotic Resistance.Microorganisms. 2022 Nov 29;10(12):2362. doi: 10.3390/microorganisms10122362. Microorganisms. 2022. PMID: 36557614 Free PMC article. Review.
-
Flux Sampling in Genome-scale Metabolic Modeling of Microbial Communities.bioRxiv [Preprint]. 2023 Apr 20:2023.04.18.537368. doi: 10.1101/2023.04.18.537368. bioRxiv. 2023. Update in: BMC Bioinformatics. 2024 Jan 29;25(1):45. doi: 10.1186/s12859-024-05655-3. PMID: 37197028 Free PMC article. Updated. Preprint.
-
Genome-Scale Metabolic Models in Fungal Pathogens: Past, Present, and Future.Int J Mol Sci. 2024 Oct 9;25(19):10852. doi: 10.3390/ijms251910852. Int J Mol Sci. 2024. PMID: 39409179 Free PMC article. Review.
-
Genome-scale model of Rothia mucilaginosa predicts gene essentialities and reveals metabolic capabilities.Microbiol Spectr. 2024 Jun 4;12(6):e0400623. doi: 10.1128/spectrum.04006-23. Epub 2024 Apr 23. Microbiol Spectr. 2024. PMID: 38652457 Free PMC article.
References
-
- AbuOun M., Suthers P. F., Jones G. I., Carter B. R., Saunders M. P., Maranas C. D., et al. . (2009). Genome scale reconstruction of a salmonella metabolic model: Comparison of similarity and differences with a commensal Escherichia coli strain. J. Biol. Chem. 284, 29480–29488. 10.1074/jbc.M109.005868 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources

