Residue L193P Mutant of RpoS Affects Its Activity During Biofilm Formation in Salmonella Pullorum
- PMID: 33251260
- PMCID: PMC7674402
- DOI: 10.3389/fvets.2020.571361
Residue L193P Mutant of RpoS Affects Its Activity During Biofilm Formation in Salmonella Pullorum
Abstract
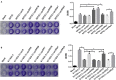
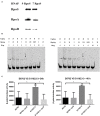
The role of alternative sigma factor RpoS in regulating biofilm formation may differ in various Salmonella Pullorum strains. In this study, the biofilm-forming ability of two Salmonella Pullorum strains S6702 and S11923-3 were compared. The biofilm forming ability of S11923-3 was much stronger than that of S6702. After knocking out the rpoS gene, S11923-3ΔrpoS had significantly reduced biofilm while S6702ΔrpoS demonstrated similar biofilm compared with each parent strain. The analysis of RpoS sequences indicated two amino acid substitutions (L193P and R293C) between S6702 and S11923-3 RpoS. A complementation study confirmed that the expression of S11923-3 RpoS rather than S6702 RpoS could restore the biofilm-forming ability of ΔrpoS strains and the L193P mutation contributed to the restoration of the biofilm-forming ability. Further study indicated that RpoS with the L193P mutant had significantly improved expression level and binding activity to RNAP and csgD gene promoter, which increased the efficacy of the csgD gene promoter and biofilm-forming ability. Therefore, the L193P mutation of RpoS is critical for stronger biofilm formation of Salmonella Pullorum.
Keywords: Salmonella pullorum; biofilm; mutation; promoter; rpoS.
Copyright © 2020 Feng, El Hag, Qin, Du, Chen and Peng.
Figures


Similar articles
-
[Identification of rpoE gene associated with biofilm formation of Salmonella pullorum].Wei Sheng Wu Xue Bao. 2015 Feb 4;55(2):156-63. Wei Sheng Wu Xue Bao. 2015. PMID: 25958695 Chinese.
-
Identification of genes responsible for biofilm formation or virulence in Salmonella enterica serovar pullorum.Avian Dis. 2012 Mar;56(1):134-43. doi: 10.1637/9806-052411-Reg.1. Avian Dis. 2012. PMID: 22545539
-
Phage insertion in mlrA and variations in rpoS limit curli expression and biofilm formation in Escherichia coli serotype O157: H7.Microbiology (Reading). 2013 Aug;159(Pt 8):1586-1596. doi: 10.1099/mic.0.066118-0. Epub 2013 Jun 6. Microbiology (Reading). 2013. PMID: 23744902
-
Mechanistic and bibliometric insights into RpoS-mediated biofilm regulation and its strategic role in food safety applications.Crit Rev Food Sci Nutr. 2025 Jan 29:1-15. doi: 10.1080/10408398.2025.2458755. Online ahead of print. Crit Rev Food Sci Nutr. 2025. PMID: 39879107 Review.
-
Recent advances in the characterization of Crl, the unconventional activator of the stress sigma factor σS/RpoS.Biomol Concepts. 2016 Jun 1;7(3):197-204. doi: 10.1515/bmc-2016-0006. Biomol Concepts. 2016. PMID: 27180360 Review.
Cited by
-
Trans-cinnamaldehyde nanoemulsion reduces Salmonella Enteritidis biofilm on steel and plastic surfaces and downregulates expression of biofilm associated genes.Poult Sci. 2025 May;104(5):105086. doi: 10.1016/j.psj.2025.105086. Epub 2025 Mar 22. Poult Sci. 2025. PMID: 40168703 Free PMC article.
-
Salmonella stress response systems as targets for anti-virulence strategies.BMC Microbiol. 2025 Jul 2;25(1):378. doi: 10.1186/s12866-025-04003-6. BMC Microbiol. 2025. PMID: 40596824 Free PMC article.
References
LinkOut - more resources
Full Text Sources

