Identification of hsa-miR-1275 as a Novel Biomarker Targeting MECP2 for Human Epilepsy of Unknown Etiology
- PMID: 33251277
- PMCID: PMC7677659
- DOI: 10.1016/j.omtm.2020.10.005
Identification of hsa-miR-1275 as a Novel Biomarker Targeting MECP2 for Human Epilepsy of Unknown Etiology
Abstract
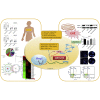
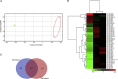
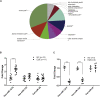
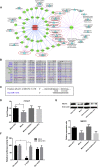
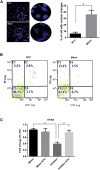
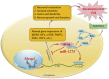
Epilepsy affects around 70 million people worldwide, with a 65% rate of unknown etiology. This rate is known as epilepsy of unknown etiology (EUE). Dysregulation of microRNAs (miRNAs) is recognized to contribute to mental disorders, including epilepsy. However, miRNA dysregulation is poorly understood in EUE. Here, we conducted miRNA expression profiling of EUE by microarray technology and identified 57 pathogenic changed miRNAs with significance. The data and bioinformatic analysis results indicated that among these miRNAs, hsa-microRNA (miR)-1275 was highly associated with neurological disorders. Subsequently, new samples of serum and cerebrospinal fluid were collected for validation of hsa-miR-1275 expression by TaqMan assays. Results show that hsa-miR-1275 in serums of EUE were increased significantly, but in cerebrospinal fluid, the miRNA was decreased. Moreover, the MECP2 gene was selected as a hsa-miR-1275 target based on target prediction tools and gene ontology analysis. Validation of in vitro tests proved that MECP2 expression was specifically inhibited by hsa-miR-1275. Additionally, overexpression of hsa-miR-1275 can elevate expression of nuclear factor κB (NF-κB) and promote cell apoptosis. Taken together, hsa-miR-1275 might represent a novel biomarker targeting MECP2 for human EUE.
Keywords: MECP2; epilepsy of unknown etiology; hsa-miR-1275; hsa-miR-132.
© 2020 The Author(s).
Figures
Similar articles
-
SOX11 identified by target gene evaluation of miRNAs differentially expressed in focal and non-focal brain tissue of therapy-resistant epilepsy patients.Neurobiol Dis. 2015 May;77:127-40. doi: 10.1016/j.nbd.2015.02.025. Epub 2015 Mar 10. Neurobiol Dis. 2015. PMID: 25766675 Free PMC article.
-
MicroRNA profiling reveals dysregulated microRNAs and their target gene regulatory networks in cemento-ossifying fibroma.J Oral Pathol Med. 2018 Jan;47(1):78-85. doi: 10.1111/jop.12650. Epub 2017 Nov 1. J Oral Pathol Med. 2018. PMID: 29032608
-
Peripheral whole blood microRNA alterations in major depression and bipolar disorder.J Affect Disord. 2016 Aug;200:250-8. doi: 10.1016/j.jad.2016.04.021. Epub 2016 Apr 20. J Affect Disord. 2016. PMID: 27152760
-
Dysregulated Serum MiRNA Profile and Promising Biomarkers in Dengue-infected Patients.Int J Med Sci. 2016 Feb 18;13(3):195-205. doi: 10.7150/ijms.13996. eCollection 2016. Int J Med Sci. 2016. PMID: 26941580 Free PMC article.
-
Serum MicroRNA-4521 is a Potential Biomarker for Focal Cortical Dysplasia with Refractory Epilepsy.Neurochem Res. 2016 Apr;41(4):905-12. doi: 10.1007/s11064-015-1773-0. Epub 2015 Dec 8. Neurochem Res. 2016. PMID: 26645999
Cited by
-
Role and Dysregulation of miRNA in Patients with Parkinson's Disease.Int J Mol Sci. 2022 Dec 31;24(1):712. doi: 10.3390/ijms24010712. Int J Mol Sci. 2022. PMID: 36614153 Free PMC article.
-
Unveiling Molecular Dynamics of MeCp2, CDKL5 and BDNF in the Hippocampus of Individuals With Intractable Mesial Temporal Lobe Epilepsy.J Cell Mol Med. 2025 Feb;29(3):e70373. doi: 10.1111/jcmm.70373. J Cell Mol Med. 2025. PMID: 39888294 Free PMC article.
-
Aberrant expression of miRNAs in epilepsy.Mol Biol Rep. 2022 Jun;49(6):5057-5074. doi: 10.1007/s11033-022-07188-5. Epub 2022 Jan 28. Mol Biol Rep. 2022. PMID: 35088379 Free PMC article. Review.
-
MicroRNAs as Potential Biomarkers of Post-Traumatic Epileptogenesis: A Systematic Review.Int J Mol Sci. 2023 Oct 19;24(20):15366. doi: 10.3390/ijms242015366. Int J Mol Sci. 2023. PMID: 37895044 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

