High affinity of host human microRNAs to SARS-CoV-2 genome: An in silico analysis
- PMID: 33251388
- PMCID: PMC7680021
- DOI: 10.1016/j.ncrna.2020.11.005
High affinity of host human microRNAs to SARS-CoV-2 genome: An in silico analysis
Abstract
Background: Coronavirus disease 2019 (COVID-19) caused by a novel betacoronavirus named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has attracted top health concerns worldwide within a few months after its appearance. Since viruses are highly dependent on the host small RNAs (microRNAs) for their replication and propagation, in this study, top miRNAs targeting SARS-CoV-2 genome and top miRNAs targeting differentially expressed genes (DEGs) in lungs of patients infected with SARS-CoV-2, were predicted.
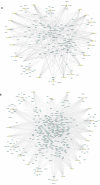
Methods: All human mature miRNA sequences were acquired from miRBase database. MiRanda tool was used to predict the potential human miRNA binding sites on the SARS-CoV-2 genome. EdgeR identified differentially expressed genes (DEGs) in response to SARS-CoV-2 infection from GEO147507 data. Gene Set Enrichment Analysis (GSEA) and DEGs annotation analysis were performed using ToppGene and Metascape tools.
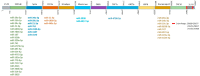
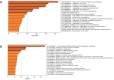
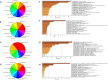
Results: 160 miRNAs with a perfect matching in the seed region were identified. Among them, there was 15 miRNAs with more than three binding sites and 12 miRNAs with a free energy binding of -29 kCal/Mol. MiR-29 family had the most binding sites (11 sites) on the SARS-CoV-2 genome. MiR-21 occupied four binding sites and was among the top miRNAs that targeted up-regulated DEGs. In addition to miR-21, miR-16, let-7b, let-7e, and miR-146a were the top miRNAs targeting DEGs.
Conclusion: Collectively, more experimental studies especially miRNA-based studies are needed to explore detailed molecular mechanisms of SARS-CoV-2 infection. Moreover, the role of DEGs including STAT1, CCND1, CXCL-10, and MAPKAPK2 in SARS-CoV-2 should be investigated to identify the similarities and differences between SARS-CoV-2 and other respiratory viruses.
Keywords: COVID-19; SARS-CoV-2; miR-21; miR-29; microRNA.
© 2020 Production and hosting by Elsevier B.V. on behalf of KeAi Communications Co., Ltd.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
Similar articles
-
A new insight into sex-specific non-coding RNAs and networks in response to SARS-CoV-2.Infect Genet Evol. 2022 Jan;97:105195. doi: 10.1016/j.meegid.2021.105195. Epub 2021 Dec 23. Infect Genet Evol. 2022. PMID: 34954105 Free PMC article.
-
Computational Analysis of Targeting SARS-CoV-2, Viral Entry Proteins ACE2 and TMPRSS2, and Interferon Genes by Host MicroRNAs.Genes (Basel). 2020 Nov 16;11(11):1354. doi: 10.3390/genes11111354. Genes (Basel). 2020. PMID: 33207533 Free PMC article.
-
IFI44 is an immune evasion biomarker for SARS-CoV-2 and Staphylococcus aureus infection in patients with RA.Front Immunol. 2022 Sep 15;13:1013322. doi: 10.3389/fimmu.2022.1013322. eCollection 2022. Front Immunol. 2022. PMID: 36189314 Free PMC article.
-
Alterations in Circulating miRNA Levels after Infection with SARS-CoV-2 Could Contribute to the Development of Cardiovascular Diseases: What We Know So Far.Int J Mol Sci. 2023 Jan 25;24(3):2380. doi: 10.3390/ijms24032380. Int J Mol Sci. 2023. PMID: 36768701 Free PMC article. Review.
-
The role of microRNAs in modulating SARS-CoV-2 infection in human cells: a systematic review.Infect Genet Evol. 2021 Jul;91:104832. doi: 10.1016/j.meegid.2021.104832. Epub 2021 Apr 1. Infect Genet Evol. 2021. PMID: 33812037 Free PMC article.
Cited by
-
Multiple functions of stress granules in viral infection at a glance.Front Microbiol. 2023 Mar 1;14:1138864. doi: 10.3389/fmicb.2023.1138864. eCollection 2023. Front Microbiol. 2023. PMID: 36937261 Free PMC article. Review.
-
miRNA expression in COVID-19.Gene Rep. 2022 Sep;28:101641. doi: 10.1016/j.genrep.2022.101641. Epub 2022 Jul 16. Gene Rep. 2022. PMID: 35875722 Free PMC article. Review.
-
Non-coding RNA in SARS-CoV-2: Progress toward therapeutic significance.Int J Biol Macromol. 2022 Dec 1;222(Pt A):1538-1550. doi: 10.1016/j.ijbiomac.2022.09.105. Epub 2022 Sep 22. Int J Biol Macromol. 2022. Retraction in: Int J Biol Macromol. 2025 Mar;293:138936. doi: 10.1016/j.ijbiomac.2024.138936. PMID: 36152703 Free PMC article. Retracted. Review.
-
Computational Prediction of RNA-RNA Interactions between Small RNA Tracks from Betacoronavirus Nonstructural Protein 3 and Neurotrophin Genes during Infection of an Epithelial Lung Cancer Cell Line: Potential Role of Novel Small Regulatory RNA.Viruses. 2023 Jul 28;15(8):1647. doi: 10.3390/v15081647. Viruses. 2023. PMID: 37631989 Free PMC article.
-
Epigenetic underpinnings of inflammation: Connecting the dots between pulmonary diseases, lung cancer and COVID-19.Semin Cancer Biol. 2022 Aug;83:384-398. doi: 10.1016/j.semcancer.2021.01.003. Epub 2021 Jan 20. Semin Cancer Biol. 2022. PMID: 33484868 Free PMC article. Review.
References
-
- Lee N., Hui D., Wu A., Chan P., Cameron P., Joynt G.M., Ahuja A., Yung M.Y., Leung C. A major outbreak of severe acute respiratory syndrome in Hong Kong. KJNEJoM. 2003;348(20):1986–1994. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

