The White-Spotted Bamboo Shark Genome Reveals Chromosome Rearrangements and Fast-Evolving Immune Genes of Cartilaginous Fish
- PMID: 33251490
- PMCID: PMC7677710
- DOI: 10.1016/j.isci.2020.101754
The White-Spotted Bamboo Shark Genome Reveals Chromosome Rearrangements and Fast-Evolving Immune Genes of Cartilaginous Fish
Abstract
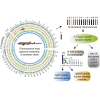
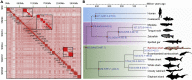
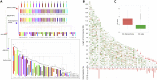
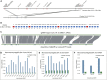
Chondrichthyan (cartilaginous fish) occupies a key phylogenetic position and is important for investigating evolutionary processes of vertebrates. However, limited whole genomes impede our in-depth knowledge of important issues such as chromosome evolution and immunity. Here, we report the chromosome-level genome of white-spotted bamboo shark. Combing it with other shark genomes, we reconstructed 16 ancestral chromosomes of bamboo shark and illustrate a dynamic chromosome rearrangement process. We found that genes on 13 fast-evolving chromosomes can be enriched in immune-related pathways. And two chromosomes contain important genes that can be used to develop single-chain antibodies, which were shown to have high affinity to human disease markers by using enzyme-linked immunosorbent assay. We also found three bone formation-related genes were lost due to chromosome rearrangements. Our study highlights the importance of chromosome rearrangements, providing resources for understanding of cartilaginous fish diversification and potential application of single-chain antibodies.
Keywords: Biological Sciences; Evolutionary Biology; Genetics; Genomics; Phylogenetics.
© 2020 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
The whale shark genome reveals patterns of vertebrate gene family evolution.Elife. 2021 Aug 19;10:e65394. doi: 10.7554/eLife.65394. Elife. 2021. PMID: 34409936 Free PMC article.
-
Elephant shark sequence reveals unique insights into the evolutionary history of vertebrate genes: A comparative analysis of the protocadherin cluster.Proc Natl Acad Sci U S A. 2008 Mar 11;105(10):3819-24. doi: 10.1073/pnas.0800398105. Epub 2008 Mar 4. Proc Natl Acad Sci U S A. 2008. PMID: 18319338 Free PMC article.
-
Characterization of Gonadotropin-Releasing Hormone (GnRH) Genes From Cartilaginous Fish: Evolutionary Perspectives.Front Neurosci. 2018 Sep 6;12:607. doi: 10.3389/fnins.2018.00607. eCollection 2018. Front Neurosci. 2018. PMID: 30237760 Free PMC article.
-
Alternative adaptive immunity strategies: coelacanth, cod and shark immunity.Mol Immunol. 2016 Jan;69:157-69. doi: 10.1016/j.molimm.2015.09.003. Epub 2015 Sep 28. Mol Immunol. 2016. PMID: 26423359 Review.
-
Shark New Antigen Receptor (IgNAR): Structure, Characteristics and Potential Biomedical Applications.Cells. 2021 May 8;10(5):1140. doi: 10.3390/cells10051140. Cells. 2021. PMID: 34066890 Free PMC article. Review.
Cited by
-
Insights into chromosomal evolution and sex determination of Pseudobagrus ussuriensis (Bagridae, Siluriformes) based on a chromosome-level genome.DNA Res. 2022 Jun 25;29(4):dsac028. doi: 10.1093/dnares/dsac028. DNA Res. 2022. PMID: 35861402 Free PMC article.
-
Comparative Genomics Reveal Phylogenetic Relationship and Chromosomal Evolutionary Events of Eight Cervidae Species.Animals (Basel). 2024 Mar 30;14(7):1063. doi: 10.3390/ani14071063. Animals (Basel). 2024. PMID: 38612302 Free PMC article.
-
Characteristics of the spiny dogfish (Squalus acanthias) nuclear genome.G3 (Bethesda). 2023 Aug 30;13(9):jkad146. doi: 10.1093/g3journal/jkad146. G3 (Bethesda). 2023. PMID: 37395764 Free PMC article.
-
A Highly Complex, MHC-Linked, 350 Million-Year-Old Shark Nonclassical Class I Lineage.J Immunol. 2021 Aug 1;207(3):824-836. doi: 10.4049/jimmunol.2000851. Epub 2021 Jul 23. J Immunol. 2021. PMID: 34301841 Free PMC article.
-
Insights into the nuclear and mitochondrial genome of the Lemon shark Negaprion brevirostris using low-coverage sequencing: Genome size, repetitive elements, mitochondrial genome, and phylogenetic placement.Gene. 2024 Feb 5;894:147939. doi: 10.1016/j.gene.2023.147939. Epub 2023 Oct 29. Gene. 2024. PMID: 38572145 Free PMC article.
References
-
- Alexander A.B., Parkinson L.A., Grant K.R., Carlson E., Campbell T.W. The hemic response of white-spotted bamboo sharks (Chiloscyllium plagiosum) with inflammatory disease. Z. Biol. 2016;35:251–259. - PubMed
-
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. - PubMed
-
- Burnstock G. Purinergic signalling: its unpopular beginning, its acceptance and its exciting future. Bioessays. 2012;34:218–225. - PubMed
-
- Cao Y., Janke A., Waddell P.J., Westerman M., Takenaka O., Murata S., Okada N., Pääbo S., Hasegawa M. Conflict among individual mitochondrial proteins in resolving the phylogeny of eutherian orders. J. Mol. Evol. 1998;47:307–322. - PubMed
LinkOut - more resources
Full Text Sources

