Screening of Methylation Gene Sites as Prognostic Signature in Lung Adenocarcinoma
- PMID: 33251775
- PMCID: PMC7700873
- DOI: 10.3349/ymj.2020.61.12.1013
Screening of Methylation Gene Sites as Prognostic Signature in Lung Adenocarcinoma
Abstract
Purpose: Most lung adenocarcinoma (LUAD) patients are diagnosed at the advanced stage and have poor prognosis. DNA methylation plays an important role in the prognosis prediction of cancers. The objective of this study was to identify new DNA methylation sites as biomarkers for LUAD prognosis.
Materials and methods: We downloaded DNA methylation data from The Cancer Genome Atlas data portal. Cox proportional hazard regression model and random survival forest algorithm were applied to identify the DNA-methylation sites. Methylation of sites were validated in the Gene Expression Omnibus cohorts. Function annotation were done to explore the biological function of DNA methylated sites signature.
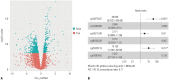
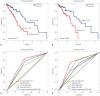
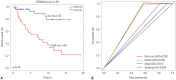
Results: Six DNA methylation sites were identified as prognosis signature. The signature yielded acceptable discrimination between the high-risk group and low-risk group. The discrimination effect of this DNA methylation signature for the OS was obvious, with a median OS of 21.89 months vs. 17.74 months for high-risk vs. low-risk groups. This prognostic prediction model was validated by the test group and GEO dataset. The predictive survival value was higher for the prognostic prediction model than that for the tumor node metastasis stage. Adjuvant hemotherapy could not affect the prediction of the signature. Functional analysis indicated that these signature genes were involved in protein binding and cytoplasm.
Conclusion: We identified the prognostic signature for LUAD by combining six DNA methylation sites. This could service as potential robust and specificity signature in the prognosis prediction of LUAD.
Keywords: Lung adenocarcinoma; TCGA; methylation; prognosis signature.
© Copyright: Yonsei University College of Medicine 2020.
Conflict of interest statement
The authors have no potential conflicts of interest to disclose.
Figures


Similar articles
-
A large cohort study identifying a novel prognosis prediction model for lung adenocarcinoma through machine learning strategies.BMC Cancer. 2019 Sep 5;19(1):886. doi: 10.1186/s12885-019-6101-7. BMC Cancer. 2019. PMID: 31488089 Free PMC article.
-
Development and validation of a robust immune-related prognostic signature in early-stage lung adenocarcinoma.J Transl Med. 2020 Oct 7;18(1):380. doi: 10.1186/s12967-020-02545-z. J Transl Med. 2020. PMID: 33028329 Free PMC article.
-
A methylation-related lncRNA-based prediction model in lung adenocarcinomas.Clin Respir J. 2024 Aug;18(8):e13753. doi: 10.1111/crj.13753. Clin Respir J. 2024. PMID: 39187946 Free PMC article.
-
Identification of prognostic values of the transcription factor-CpG-gene triplets in lung adenocarcinoma: A narrative review.Medicine (Baltimore). 2022 Dec 16;101(50):e32045. doi: 10.1097/MD.0000000000032045. Medicine (Baltimore). 2022. PMID: 36550923 Free PMC article. Review.
-
Methylation in Lung Cancer: A Brief Review.Methods Mol Biol. 2020;2204:91-97. doi: 10.1007/978-1-0716-0904-0_8. Methods Mol Biol. 2020. PMID: 32710317 Review.
Cited by
-
A differentially-methylated-region signature predicts the recurrence risk for patients with early stage lung adenocarcinoma.Aging (Albany NY). 2024 Nov 18;16(21):13323-13339. doi: 10.18632/aging.206139. Epub 2024 Nov 18. Aging (Albany NY). 2024. PMID: 39560475 Free PMC article.
-
A gene signature driven by abnormally methylated DEGs was developed for TP53 wild-type ovarian cancer samples by integrative omics analysis of DNA methylation and gene expression data.Ann Transl Med. 2023 Jan 15;11(1):20. doi: 10.21037/atm-22-5764. Ann Transl Med. 2023. PMID: 36760264 Free PMC article.
-
Current Adenosinergic Therapies: What Do Cancer Cells Stand to Gain and Lose?Int J Mol Sci. 2021 Nov 22;22(22):12569. doi: 10.3390/ijms222212569. Int J Mol Sci. 2021. PMID: 34830449 Free PMC article. Review.
-
NUF2 Expression Promotes Lung Adenocarcinoma Progression and Is Associated With Poor Prognosis.Front Oncol. 2022 Jun 23;12:795971. doi: 10.3389/fonc.2022.795971. eCollection 2022. Front Oncol. 2022. PMID: 35814368 Free PMC article.
-
Identification and Validation of a Prognostic Signature Based on Methylation Profiles and Methylation-Driven Gene DAB2 as a Prognostic Biomarker in Differentiated Thyroid Carcinoma.Dis Markers. 2022 Sep 17;2022:1686316. doi: 10.1155/2022/1686316. eCollection 2022. Dis Markers. 2022. PMID: 37223105 Free PMC article.
References
-
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. - PubMed
-
- Eberhardt WE, Stuschke M. Multimodal treatment of non-small-cell lung cancer. Lancet. 2015;386:1018–1020. - PubMed
-
- Ettinger DS, Akerley W, Borghaei H, Chang AC, Cheney RT, Chirieac LR, et al. Non-small cell lung cancer, version 2.2013. J Natl Compr Canc Netw. 2013;11:645–653. - PubMed
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

