Impact of Preanalytical Factors During Histology Processing on Section Suitability for Digital Image Analysis
- PMID: 33251977
- PMCID: PMC8091422
- DOI: 10.1177/0192623320970534
Impact of Preanalytical Factors During Histology Processing on Section Suitability for Digital Image Analysis
Abstract
Digital image analysis (DIA) is impacted by the quality of tissue staining. This study examined the influence of preanalytical variables-staining protocol design, reagent quality, section attributes, and instrumentation-on the performance of automated DIA software. Our hypotheses were that (1) staining intensity is impacted by subtle differences in protocol design, reagent quality, and section composition and that (2) identically programmed and loaded stainers will produce equivalent immunohistochemical (IHC) staining. We tested these propositions by using 1 hematoxylin and eosin stainer to process 13 formalin-fixed, paraffin-embedded (FFPE) mouse tissues and by using 3 identically programmed and loaded immunostainers to process 5 FFPE mouse tissues for 4 cell biomarkers. Digital images of stained sections acquired with a commercial whole slide scanner were analyzed by customizable algorithms incorporated into commercially available DIA software. Staining intensity as viewed qualitatively by an observer and/or quantitatively by DIA was affected by staining conditions and tissue attributes. Intrarun and inter-run IHC staining intensities were equivalent for each tissue when processed on a given stainer but varied measurably across stainers. Our data indicate that staining quality must be monitored for each method and stainer to ensure that preanalytical factors do not impact digital pathology data quality.
Keywords: digital pathology; histology process validation; image analysis; immunohistochemistry; preanalytical factors; precision; reproducibility.
Conflict of interest statement
Figures
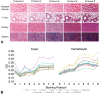
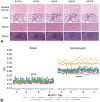
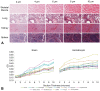

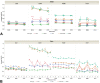






References
-
- Webster JD, Dunstan RW. Whole-slide imaging and automated image analysis: considerations and opportunities in the practice of pathology. Vet Pathol. 2014;51(1):211–223. - PubMed
-
- Potts SJ. Digital pathology in drug discovery and development: multisite integration. Drug Discov Today. 2009;14(19-20):935–941. - PubMed
-
- Gauthier BE, Gervais F, Hamm G, O’Shea D, Piton A, Schumacher VL. Toxicologic Pathology Forum: opinion on integrating innovative digital pathology tools in the regulatory framework. Toxicol Pathol. 2019;47(4):436–443. - PubMed
-
- Griffin J, Treanor D. Digital pathology in clinical use: where are we now and what is holding us back? Histopathology. 2017;70(1):134–145. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

