One-Step Homology Mediated CRISPR-Cas Editing in Zygotes for Generating Genome Edited Cattle
- PMID: 33252243
- PMCID: PMC7757693
- DOI: 10.1089/crispr.2020.0047
One-Step Homology Mediated CRISPR-Cas Editing in Zygotes for Generating Genome Edited Cattle
Abstract
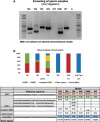
Selective breeding and genetic modification have been the cornerstone of animal agriculture. However, the current strategy of breeding animals over multiple generations to introgress novel alleles is not practical in addressing global challenges such as climate change, pandemics, and the predicted need to feed a population of 9 billion by 2050. Consequently, genome editing in zygotes to allow for seamless introgression of novel alleles is required, especially in cattle with long generation intervals. We report for the first time the use of CRISPR-Cas genome editors to introduce novel PRNP allelic variants that have been shown to provide resilience towards human prion pandemics. From one round of embryo injections, we have established six pregnancies and birth of seven edited offspring, with two founders showing >90% targeted homology-directed repair modifications. This study lays out the framework for in vitro optimization, unbiased deep-sequencing to identify editing outcomes, and generation of high frequency homology-directed repair-edited calves.
Conflict of interest statement
KP, SS, and BT serve as consultants at RenOVAte Biosciences Inc, (RBI). BT and KP are founding members of RBI. JW is employed by RBI. SP and MC are employed by Genus plc. All remaining authors declare no competing financial interests.
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

