Acetylation-mediated remodeling of the nucleolus regulates cellular acetyl-CoA responses
- PMID: 33253182
- PMCID: PMC7728262
- DOI: 10.1371/journal.pbio.3000981
Acetylation-mediated remodeling of the nucleolus regulates cellular acetyl-CoA responses
Abstract
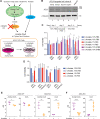
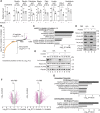
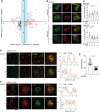
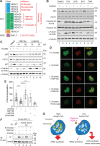
The metabolite acetyl-coenzyme A (acetyl-CoA) serves as an essential element for a wide range of cellular functions including adenosine triphosphate (ATP) production, lipid synthesis, and protein acetylation. Intracellular acetyl-CoA concentrations are associated with nutrient availability, but the mechanisms by which a cell responds to fluctuations in acetyl-CoA levels remain elusive. Here, we generate a cell system to selectively manipulate the nucleo-cytoplasmic levels of acetyl-CoA using clustered regularly interspaced short palindromic repeat (CRISPR)-mediated gene editing and acetate supplementation of the culture media. Using this system and quantitative omics analyses, we demonstrate that acetyl-CoA depletion alters the integrity of the nucleolus, impairing ribosomal RNA synthesis and evoking the ribosomal protein-dependent activation of p53. This nucleolar remodeling appears to be mediated through the class IIa histone deacetylases (HDACs). Our findings highlight acetylation-mediated control of the nucleolus as an important hub linking acetyl-CoA fluctuations to cellular stress responses.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

