Antimicrobial peptide activity is anticorrelated with lipid a leaflet affinity
- PMID: 33253275
- PMCID: PMC7703904
- DOI: 10.1371/journal.pone.0242907
Antimicrobial peptide activity is anticorrelated with lipid a leaflet affinity
Abstract
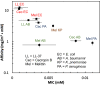
The activity of antimicrobial peptides (AMPs) has significant bacterial species bias, the mechanisms of which are not fully understood. We employed single-molecule tracking to measure the affinity of three different AMPs to hybrid supported bilayers composed of lipid A extracted from four different Gram negative bacteria and observed a strong empirical anticorrelation between the affinity of a particular AMP to a given lipid A layer and the activity of that AMP towards the bacterium from which that lipid A was extracted. This suggested that the species bias of AMP activity is directly related to AMP interactions with bacterial outer membranes, despite the fact that the mechanism of antimicrobial activity occurs at the inner membrane. The trend also suggested that the interactions between AMPs and the outer membrane lipid A (even in the absence of other components, such as lipopolysaccharides) capture effects that are relevant to the minimum inhibitory concentration.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Binding of an antimicrobial peptide to bacterial cells: Interaction with different species, strains and cellular components.Biochim Biophys Acta Biomembr. 2020 Aug 1;1862(8):183291. doi: 10.1016/j.bbamem.2020.183291. Epub 2020 Mar 28. Biochim Biophys Acta Biomembr. 2020. PMID: 32234322
-
Structural Disruptions of the Outer Membranes of Gram-Negative Bacteria by Rationally Designed Amphiphilic Antimicrobial Peptides.ACS Appl Mater Interfaces. 2021 Apr 14;13(14):16062-16074. doi: 10.1021/acsami.1c01643. Epub 2021 Apr 2. ACS Appl Mater Interfaces. 2021. PMID: 33797891
-
A Molecular Dynamics Study of Antimicrobial Peptide Interactions with the Lipopolysaccharides of the Outer Bacterial Membrane.J Membr Biol. 2022 Dec;255(6):665-675. doi: 10.1007/s00232-022-00258-6. Epub 2022 Aug 12. J Membr Biol. 2022. PMID: 35960325
-
An intimate link between antimicrobial peptide sequence diversity and binding to essential components of bacterial membranes.Biochim Biophys Acta. 2016 May;1858(5):958-70. doi: 10.1016/j.bbamem.2015.10.011. Epub 2015 Oct 21. Biochim Biophys Acta. 2016. PMID: 26498397 Review.
-
Molecular mechanisms of membrane targeting antibiotics.Biochim Biophys Acta. 2016 May;1858(5):980-7. doi: 10.1016/j.bbamem.2015.10.018. Epub 2015 Oct 26. Biochim Biophys Acta. 2016. PMID: 26514603 Review.
Cited by
-
How to Combat Gram-Negative Bacteria Using Antimicrobial Peptides: A Challenge or an Unattainable Goal?Antibiotics (Basel). 2021 Dec 7;10(12):1499. doi: 10.3390/antibiotics10121499. Antibiotics (Basel). 2021. PMID: 34943713 Free PMC article. Review.
-
Incorporation of Non-Canonical Amino Acids into Antimicrobial Peptides: Advances, Challenges, and Perspectives.Appl Environ Microbiol. 2022 Dec 13;88(23):e0161722. doi: 10.1128/aem.01617-22. Epub 2022 Nov 23. Appl Environ Microbiol. 2022. PMID: 36416555 Free PMC article. Review.
-
Solid and Liquid Surface-Supported Bacterial Membrane Mimetics as a Platform for the Functional and Structural Studies of Antimicrobials.Membranes (Basel). 2022 Sep 20;12(10):906. doi: 10.3390/membranes12100906. Membranes (Basel). 2022. PMID: 36295664 Free PMC article. Review.
-
Plasticity and Stereotypic Rewiring of the Transcriptome Upon Bacterial Evolution of Antibiotic Resistance.Mol Biol Evol. 2023 Feb 3;40(2):msad020. doi: 10.1093/molbev/msad020. Mol Biol Evol. 2023. PMID: 36718533 Free PMC article.
-
Chaotropic Agent-Assisted Supported Lipid Bilayer Formation.Langmuir. 2024 Oct 1;40(39):20629-20639. doi: 10.1021/acs.langmuir.4c02543. Epub 2024 Sep 17. Langmuir. 2024. PMID: 39285818 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

