A population genomic unveiling of a new cryptic mosquito taxon within the malaria-transmitting Anopheles gambiae complex
- PMID: 33253481
- PMCID: PMC7858241
- DOI: 10.1111/mec.15756
A population genomic unveiling of a new cryptic mosquito taxon within the malaria-transmitting Anopheles gambiae complex
Abstract
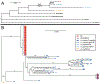
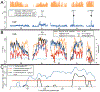
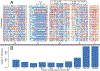
The Anopheles gambiae complex consists of multiple morphologically indistinguishable mosquito species including the most important vectors of the malaria parasite Plasmodium falciparum in sub-Saharan Africa. Nine cryptic species have been described so far within the complex. The ecological, immunological and reproductive differences among these species will critically impact population responses to disease control strategies and environmental changes. Here, we examine whole-genome sequencing data from a longitudinal study of putative A. coluzzii in western Burkina Faso. Surprisingly, many specimens are genetically divergent from A. coluzzii and all other Anopheles species and represent a new taxon, here designated Anopheles TENGRELA (AT). Population genetic analysis suggests that the cryptic GOUNDRY subgroup, previously collected as larvae in central Burkina Faso, represents an admixed population descended from both A. coluzzii and AT. AT harbours low nucleotide diversity except for the 2La inversion polymorphism which is maintained by overdominance. It shows numerous fixed differences with A. coluzzii concentrated in several regions reflecting selective sweeps, but the two taxa are identical at standard diagnostic loci used for taxon identification, and thus, AT may often go unnoticed. We present an amplicon-based genotyping assay for identifying AT which could be usefully applied to numerous existing samples. Misidentified cryptic taxa could seriously confound ongoing studies of Anopheles ecology and evolution in western Africa, including phenotypic and genotypic surveys of insecticide resistance. Reproductive barriers between cryptic species may also complicate novel vector control efforts, for example gene drives, and hinder predictions about evolutionary dynamics of Anopheles and Plasmodium.
Keywords: Anopheles; admixture; cryptic taxa; reproductive barrier; selective sweep; vector.
© 2020 John Wiley & Sons Ltd.
Figures
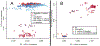

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

