WWOX Loss of Function in Neurodevelopmental and Neurodegenerative Disorders
- PMID: 33255508
- PMCID: PMC7727818
- DOI: 10.3390/ijms21238922
WWOX Loss of Function in Neurodevelopmental and Neurodegenerative Disorders
Abstract
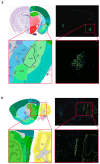
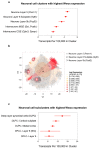
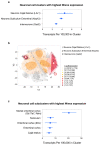
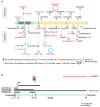
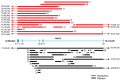
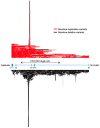
The WWOX gene was initially discovered as a putative tumor suppressor. More recently, its association with multiple central nervous system (CNS) pathologies has been recognized. WWOX biallelic germline pathogenic variants have been implicated in spinocerebellar ataxia type 12 (SCAR12; MIM:614322) and in early infantile epileptic encephalopathy (EIEE28; MIM:616211). WWOX germline copy number variants have also been associated with autism spectrum disorder (ASD). All identified germline genomic variants lead to partial or complete loss of WWOX function. Importantly, large-scale genome-wide association studies have also identified WWOX as a risk gene for common neurodegenerative conditions such as Alzheimer's disease (AD) and multiple sclerosis (MS). Thus, the spectrum of CNS disorders associated with WWOX is broad and heterogeneous, and there is little understanding of potential mechanisms at play. Exploration of gene expression databases indicates that WWOX expression is comparatively higher in the human cerebellar cortex than in other CNS structures. However, RNA in-situ hybridization data from the Allen Mouse Brain Atlas show that specific regions of the basolateral amygdala (BLA), the medial entorhinal cortex (EC), and deep layers of the isocortex can be singled out as brain regions with specific higher levels of Wwox expression. These observations are in close agreement with single-cell RNA-seq data which indicate that neurons from the medial entorhinal cortex, Layer 5 from the frontal cortex as well as GABAergic basket cells and granule cells from cerebellar cortex are the specific neuronal subtypes that display the highest Wwox expression levels. Importantly, the brain regions and cell types in which WWOX is most abundantly expressed, such as the EC and BLA, are intimately linked to pathologies and syndromic conditions in turn associated with this gene, such as epilepsy, intellectual disability, ASD, and AD. Higher Wwox expression in interneurons and granule cells from cerebellum points to a direct link to the described cerebellar ataxia in cases of WWOX loss of function. We now know that total or partial impairment of WWOX function results in a wide and heterogeneous variety of neurodegenerative conditions for which the specific molecular mechanisms remain to be deciphered. Nevertheless, these observations indicate an important functional role for WWOX in normal development and function of the CNS. Evidence also indicates that disruption of WWOX expression at the gene or protein level in CNS has significant deleterious consequences.
Keywords: ADHD; Alzheimer’s disease; WOREE; WWOX; autism; epileptic encephalopathy; intellectual disability; multiple sclerosis; neurodegeneration; spinocerebellar ataxia.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Bednarek A.K., Laflin K.J., Daniel R.L., Liao Q., Hawkins K.A., Aldaz C.M. WWOX, a novel WW domain-containing protein mapping to human chromosome 16q23.3-24.1, a region frequently affected in breast cancer. Cancer Res. 2000;60:2140–2145. - PubMed
-
- Bednarek A.K., Keck-Waggoner C.L., Daniel R.L., Laflin K.J., Bergsagel P.L., Kiguchi K., Brenner A.J., Aldaz C.M. WWOX, the FRA16D gene, behaves as a suppressor of tumor growth. Cancer Res. 2001;61:8068–8073. - PubMed
-
- Mallaret M., Synofzik M., Lee J., Sagum C.A., Mahajnah M., Sharkia R., Drouot N., Renaud M., Klein F.A., Anheim M., et al. The tumour suppressor gene WWOX is mutated in autosomal recessive cerebellar ataxia with epilepsy and mental retardation. Brain. 2014;137:411–419. doi: 10.1093/brain/awt338. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

