In Silico Design of a Peptide Receptor for Dopamine Recognition
- PMID: 33255517
- PMCID: PMC7727804
- DOI: 10.3390/molecules25235509
In Silico Design of a Peptide Receptor for Dopamine Recognition
Abstract
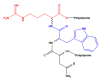
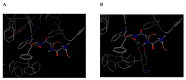
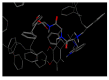
Dopamine (DA) is an important neurotransmitter with a fundamental role in regulatory functions related to the central, peripheral, renal, and hormonal nervous systems. Dopaminergic neurotransmission dysfunctions are commonly associated with several diseases; thus, in situ quantification of DA is a major challenge. To achieve this goal, enzyme-based biosensors have been employed for substrate recognition in the past. However, due to their sensitivity to changes in temperature and pH levels, new peptide bioreceptors have been developed. Therefore, in this study, four bioreceptors were designed in silico to exhibit a higher affinity for DA than the DA transporters (DATs). The design was based on the hot spots of the active sites of crystallized enzyme structures that are physiologically related to DA. The affinities between the chosen targets and designed bioreceptors were calculated using AutoDock Vina. Additionally, the binding free energy, ∆G, of the dopamine-4xp1 complex was calculated by molecular dynamics (MD). This value presented a direct relationship with the E_refine value obtained from molecular docking based on the ∆G functions obtained from MOE of the promising bioreceptors. The control variables in the design were amino acids, bond type, steric volume, stereochemistry, affinity, and interaction distances. As part of the results, three out of the four bioreceptor candidates presented promising values in terms of DA affinity and distance.
Keywords: bioreceptor; dopamine; in silico; molecular docking; molecular dynamics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
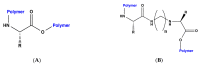







Similar articles
-
An in silico design method of a peptide bioreceptor for cortisol using molecular modelling techniques.Sci Rep. 2024 Sep 27;14(1):22325. doi: 10.1038/s41598-024-73044-0. Sci Rep. 2024. PMID: 39333310 Free PMC article.
-
Human dopamine transporter: the first implementation of a combined in silico/in vitro approach revealing the substrate and inhibitor specificities.J Biomol Struct Dyn. 2019 Feb;37(2):291-306. doi: 10.1080/07391102.2018.1426044. Epub 2018 Jan 26. J Biomol Struct Dyn. 2019. PMID: 29334320
-
Structural insights into pharmacophore-assisted in silico identification of protein-protein interaction inhibitors for inhibition of human toll-like receptor 4 - myeloid differentiation factor-2 (hTLR4-MD-2) complex.J Biomol Struct Dyn. 2019 May;37(8):1968-1991. doi: 10.1080/07391102.2018.1474804. Epub 2018 May 29. J Biomol Struct Dyn. 2019. PMID: 29842849
-
Design, synthesis and preliminary evaluation of dopamine-amino acid conjugates as potential D1 dopaminergic modulators.Eur J Med Chem. 2016 Nov 29;124:435-444. doi: 10.1016/j.ejmech.2016.08.051. Epub 2016 Aug 24. Eur J Med Chem. 2016. PMID: 27597419
-
Molecular Mechanism of Dopamine Transport by Human Dopamine Transporter.Structure. 2015 Nov 3;23(11):2171-81. doi: 10.1016/j.str.2015.09.001. Epub 2015 Oct 15. Structure. 2015. PMID: 26481814 Free PMC article.
References
-
- Rozet E., Morello R., LeComte F., Martin G., Chiap P., Crommen J., Boos K., Hubert P. Performances of a multidimensional on-line SPE-LC-ECD method for the determination of three major catecholamines in native human urine: Validation, risk and uncertainty assessments. J. Chromatogr. B. 2006;844:251–260. doi: 10.1016/j.jchromb.2006.07.060. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

