Extracellular Vesicles and Post-Translational Protein Deimination Signatures in Mollusca-The Blue Mussel (Mytilus edulis), Soft Shell Clam (Mya arenaria), Eastern Oyster (Crassostrea virginica) and Atlantic Jacknife Clam (Ensis leei)
- PMID: 33255637
- PMCID: PMC7760292
- DOI: 10.3390/biology9120416
Extracellular Vesicles and Post-Translational Protein Deimination Signatures in Mollusca-The Blue Mussel (Mytilus edulis), Soft Shell Clam (Mya arenaria), Eastern Oyster (Crassostrea virginica) and Atlantic Jacknife Clam (Ensis leei)
Abstract
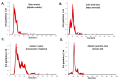
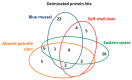
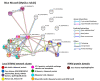
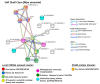
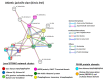
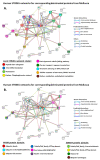
Oysters and clams are important for food security and of commercial value worldwide. They are affected by anthropogenic changes and opportunistic pathogens and can be indicators of changes in ocean environments. Therefore, studies into biomarker discovery are of considerable value. This study aimed at assessing extracellular vesicle (EV) signatures and post-translational protein deimination profiles of hemolymph from four commercially valuable Mollusca species, the blue mussel (Mytilus edulis), soft shell clam (Mya arenaria), Eastern oyster (Crassostrea virginica), and Atlantic jacknife clam (Ensis leei). EVs form part of cellular communication by transporting protein and genetic cargo and play roles in immunity and host-pathogen interactions. Protein deimination is a post-translational modification caused by peptidylarginine deiminases (PADs), and can facilitate protein moonlighting in health and disease. The current study identified hemolymph-EV profiles in the four Mollusca species, revealing some species differences. Deiminated protein candidates differed in hemolymph between the species, with some common targets between all four species (e.g., histone H3 and H4, actin, and GAPDH), while other hits were species-specific; in blue mussel these included heavy metal binding protein, heat shock proteins 60 and 90, 2-phospho-D-glycerate hydrolyase, GTP cyclohydrolase feedback regulatory protein, sodium/potassium-transporting ATPase, and fibrinogen domain containing protein. In soft shell clam specific deimination hits included dynein, MCM3-associated protein, and SCRN. In Eastern oyster specific deimination hits included muscle LIM protein, beta-1,3-glucan-binding protein, myosin heavy chain, thaumatin-like protein, vWFA domain-containing protein, BTB domain-containing protein, amylase, and beta-catenin. Deiminated proteins specific to Atlantic jackknife clam included nacre c1q domain-containing protein and PDZ domain-containing protein In addition, some proteins were common as deiminated targets between two or three of the Bivalvia species under study (e.g., EP protein, C1q domain containing protein, histone H2B, tubulin, elongation factor 1-alpha, dominin, extracellular superoxide dismutase). Protein interaction network analysis for the deiminated protein hits revealed major pathways relevant for immunity and metabolism, providing novel insights into post-translational regulation via deimination. The study contributes to EV characterization in diverse taxa and understanding of roles for PAD-mediated regulation of immune and metabolic pathways throughout phylogeny.
Keywords: Mollusca; clam; extracellular vesicles (EVs); immunity; metabolism; oyster; peptidylarginine deiminase (PAD); protein deimination/citrullination.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- FAO . The State of World Fisheries and Aquaculture Sustainability in Action. Food and Agriculture Organization; Rome, Italy: 2014. - DOI
-
- Harney E., Artigaud S., Le Souchu P., Miner P., Corporeau C., Essid H., Pichereau V., Nunes F.L.D. Non-additive effects of ocean acidification in combination with warming on the larval proteome of the Pacific oyster, Crassostrea Gigas. J. Proteomics. 2016;135:151–161. doi: 10.1016/j.jprot.2015.12.001. - DOI - PubMed
-
- Gutierrez J.L., Jones C.G., Strayer D.L., Iribarne O.O. Mollusks as ecosystem engineers: The role of shell production in aquatic habitats. Oikos. 2003;101:79–90. doi: 10.1034/j.1600-0706.2003.12322.x. - DOI
-
- Jordan S.J., Coakley J.M. Long-term projections of eastern oyster populations under various management scenarios. J. Shellfish Res. 2004;23:63–72.
LinkOut - more resources
Full Text Sources
Research Materials

