Epidemiology and Sequence-Based Evolutionary Analysis of Circulating Non-Polio Enteroviruses
- PMID: 33255654
- PMCID: PMC7759938
- DOI: 10.3390/microorganisms8121856
Epidemiology and Sequence-Based Evolutionary Analysis of Circulating Non-Polio Enteroviruses
Abstract
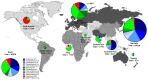
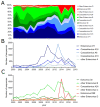
Enteroviruses (EVs) are positive-sense RNA viruses, with over 50,000 nucleotide sequences publicly available. While most human infections are typically associated with mild respiratory symptoms, several different EV types have also been associated with severe human disease, especially acute flaccid paralysis (AFP), particularly with endemic members of the EV-B species and two pandemic types-EV-A71 and EV-D68-that appear to be responsible for recent widespread outbreaks. Here we review the recent literature on the prevalence, characteristics, and circulation dynamics of different enterovirus types and combine this with an analysis of the sequence coverage of different EV types in public databases (e.g., the Virus Pathogen Resource). This evaluation reveals temporal and geographic differences in EV circulation and sequence distribution, highlighting recent EV outbreaks and revealing gaps in sequence coverage. Phylogenetic analysis of the EV genus shows the relatedness of different EV types. Recombination analysis of the EV-A species provides evidence for recombination as a mechanism of genomic diversification. The absence of broadly protective vaccines and effective antivirals makes human enteroviruses important pathogens of public health concern.
Keywords: Echovirus 30 (E30); Virus Pathogen Resource (ViPR); acute flaccid myelitis (AFM); acute flaccid paralysis (AFP); and mouth disease (HFMD); coxsackievirus A16 (CV-A16); coxsackievirus A24 (CV-A24); coxsackievirus A6 (CV-A6); enterovirus A71 (EV-A71); enterovirus D68 (EV-D68); foot; hand.
Conflict of interest statement
The authors declare no conflict of interest. The sponsors had no role in the design, execution, interpretation, or writing of the study/review.
Figures



References
-
- Pallansch M.A., Roos R.P. Enteroviruses: Polioviruses, coxsackieviruses, echoviruses, and newer enteroviruses. In: Knipe D.M., Howley P.M., Griffin D.E., Lamb R.A., Martin M.A., Roizman B., Stephen E.S., editors. Fields Virology. 4th ed. Lippincott Williams & Wilkins; Philadelphia, PA, USA: 2001. pp. 723–775.
-
- Tangermann R.H., Aylward R., Birmingham M., Horner R., Olivé J.M., Nkowane B.M., Hull H.F., Burton A. Current status of the global eradication of poliomyelitis. World Health Stat. Q. Rapp. Trimest. Stat. Sanit. Mond. 1997;50:188–194. - PubMed
-
- Shaghaghi M., Soleyman-Jahi S., Abolhassani H., Yazdani R., Azizi G., Rezaei N., Barbouche M.-R., McKinlay M.A., Aghamohammadi A. New insights into physiopathology of immunodeficiency-associated vaccine-derived poliovirus infection; systematic review of over 5 decades of data. Vaccine. 2018;36:1711–1719. doi: 10.1016/j.vaccine.2018.02.059. - DOI - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

