Metagenomics-Based Proficiency Test of Smoked Salmon Spiked with a Mock Community
- PMID: 33255715
- PMCID: PMC7760972
- DOI: 10.3390/microorganisms8121861
Metagenomics-Based Proficiency Test of Smoked Salmon Spiked with a Mock Community
Abstract
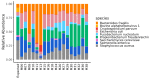
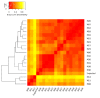
An inter-laboratory proficiency test was organized to assess the ability of participants to perform shotgun metagenomic sequencing of cold smoked salmon, experimentally spiked with a mock community composed of six bacteria, one parasite, one yeast, one DNA, and two RNA viruses. Each participant applied its in-house wet-lab workflow(s) to obtain the metagenomic dataset(s), which were then collected and analyzed using MG-RAST. A total of 27 datasets were analyzed. Sample pre-processing, DNA extraction protocol, library preparation kit, and sequencing platform, influenced the abundance of specific microorganisms of the mock community. Our results highlight that despite differences in wet-lab protocols, the reads corresponding to the mock community members spiked in the cold smoked salmon, were both detected and quantified in terms of relative abundance, in the metagenomic datasets, proving the suitability of shotgun metagenomic sequencing as a genomic tool to detect microorganisms belonging to different domains in the same food matrix. The implementation of standardized wet-lab protocols would highly facilitate the comparability of shotgun metagenomic sequencing dataset across laboratories and sectors. Moreover, there is a need for clearly defining a sequencing reads threshold, to consider pathogens as detected or undetected in a food sample.
Keywords: experimentally spiked samples; proficiency test; shotgun metagenomics; smoked salmon; wet-lab protocols.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Application of shotgun metagenomics to smoked salmon experimentally spiked: Comparison between sequencing and microbiological data using different bioinformatic approaches.Ital J Food Saf. 2019 Dec 5;8(4):8462. doi: 10.4081/ijfs.2019.8462. eCollection 2019 Dec 5. Ital J Food Saf. 2019. PMID: 31897401 Free PMC article.
-
DNA extraction protocols for animal fecal material on blood spot cards.PLoS One. 2025 May 12;20(5):e0313808. doi: 10.1371/journal.pone.0313808. eCollection 2025. PLoS One. 2025. PMID: 40354439 Free PMC article.
-
Fishing in the Soup - Pathogen Detection in Food Safety Using Metabarcoding and Metagenomic Sequencing.Front Microbiol. 2019 Aug 6;10:1805. doi: 10.3389/fmicb.2019.01805. eCollection 2019. Front Microbiol. 2019. PMID: 31447815 Free PMC article.
-
Multicenter benchmarking of short and long read wet lab protocols for clinical viral metagenomics.J Clin Virol. 2024 Aug;173:105695. doi: 10.1016/j.jcv.2024.105695. Epub 2024 May 25. J Clin Virol. 2024. PMID: 38823290
-
Surveillance of Foodborne Pathogens: Towards Diagnostic Metagenomics of Fecal Samples.Genes (Basel). 2018 Jan 4;9(1):14. doi: 10.3390/genes9010014. Genes (Basel). 2018. PMID: 29300319 Free PMC article. Review.
Cited by
-
One Health surveillance-A cross-sectoral detection, characterization, and notification of foodborne pathogens.Front Public Health. 2023 Mar 8;11:1129083. doi: 10.3389/fpubh.2023.1129083. eCollection 2023. Front Public Health. 2023. PMID: 36969662 Free PMC article.
-
Detection of parasites in food and water matrices by shotgun metagenomics: A narrative review.Food Waterborne Parasitol. 2025 Apr 21;39:e00265. doi: 10.1016/j.fawpar.2025.e00265. eCollection 2025 Jun. Food Waterborne Parasitol. 2025. PMID: 40416301 Free PMC article. Review.
-
Monitoring and preventing foodborne outbreaks: are we missing wastewater as a key data source?Ital J Food Saf. 2024 Oct 10;13(4):12725. doi: 10.4081/ijfs.2024.12725. eCollection 2024 Nov 12. Ital J Food Saf. 2024. PMID: 39749179 Free PMC article. Review.
-
Application of a strain-level shotgun metagenomics approach on food samples: resolution of the source of a Salmonella food-borne outbreak.Microb Genom. 2021 Apr;7(4):000547. doi: 10.1099/mgen.0.000547. Microb Genom. 2021. PMID: 33826490 Free PMC article.
-
Enhancing Clinical Utility: Utilization of International Standards and Guidelines for Metagenomic Sequencing in Infectious Disease Diagnosis.Int J Mol Sci. 2024 Mar 15;25(6):3333. doi: 10.3390/ijms25063333. Int J Mol Sci. 2024. PMID: 38542307 Free PMC article. Review.
References
-
- Yang X., Noyes N.R., Doster E., Martin J.N., Linke L.M., Magnuson R.J., Hua Y., Ifigenia G., Dale R.W., Kenneth L.J., et al. Use of metagenomic shotgun sequencing technology to detect foodborne pathogens within the microbiome of the beef production chain. Appl. Environ. Microbiol. 2016;82:2433–2443. doi: 10.1128/AEM.00078-16. - DOI - PMC - PubMed
-
- Larsen M.H., Dalmasso M., Ingmer H., Langsrud S., Malakauskas M., Mader A., Trond Møretrø T., Sonja Smole Mozina S.S., Rychli K., Wagner M., et al. Persistence of foodborne pathogens and their control in primary and secondary food production chains. Food Cont. 2014;44:92–109. doi: 10.1016/j.foodcont.2014.03.039. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

