Evolution of Homeologous Gene Expression in Polyploid Wheat
- PMID: 33255795
- PMCID: PMC7759873
- DOI: 10.3390/genes11121401
Evolution of Homeologous Gene Expression in Polyploid Wheat
Abstract
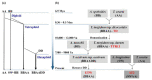
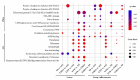
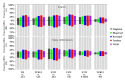
Polyploidization has played a prominent role in the evolutionary history of plants. Two recent and sequential allopolyploidization events have resulted in the formation of wheat species with different ploidies, and which provide a model to study the effects of polyploidization on the evolution of gene expression. In this study, we identified differentially expressed genes (DEGs) between four BBAA tetraploid wheats of three different ploidy backgrounds. DEGs were found to be unevenly distributed among functional categories and duplication modes. We observed more DEGs in the extracted tetraploid wheat (ETW) than in natural tetraploid wheats (TD and TTR13) as compared to a synthetic tetraploid (AT2). Furthermore, DEGs showed higher Ka/Ks ratios than those that did not show expression changes (non-DEGs) between genotypes, indicating DEGs and non-DEGs experienced different selection pressures. For A-B homeolog pairs with DEGs, most of them had only one differentially expressed copy, however, when both copies of a homeolog pair were DEGs, the A and B copies were more likely to be regulated to the same direction. Our results suggest that both cis- and inter-subgenome trans-regulatory changes are important drivers in the evolution of homeologous gene expression in polyploid wheat, with ploidy playing a significant role in the process.
Keywords: differentially expressed gene; polyploidy; transcriptome; wheat; whole genome duplication.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Transcriptome asymmetry in synthetic and natural allotetraploid wheats, revealed by RNA-sequencing.New Phytol. 2016 Feb;209(3):1264-77. doi: 10.1111/nph.13678. Epub 2015 Oct 5. New Phytol. 2016. PMID: 26436593
-
Evolution of the BBAA component of bread wheat during its history at the allohexaploid level.Plant Cell. 2014 Jul;26(7):2761-76. doi: 10.1105/tpc.114.128439. Epub 2014 Jul 2. Plant Cell. 2014. PMID: 24989045 Free PMC article.
-
Wheat in vivo RNA structure landscape reveals a prevalent role of RNA structure in modulating translational subgenome expression asymmetry.Genome Biol. 2021 Nov 30;22(1):326. doi: 10.1186/s13059-021-02549-y. Genome Biol. 2021. PMID: 34847934 Free PMC article.
-
Making the Bread: Insights from Newly Synthesized Allohexaploid Wheat.Mol Plant. 2015 Jun;8(6):847-59. doi: 10.1016/j.molp.2015.02.016. Epub 2015 Mar 5. Mol Plant. 2015. PMID: 25747845 Review.
-
Homeolog expression quantification methods for allopolyploids.Brief Bioinform. 2020 Mar 23;21(2):395-407. doi: 10.1093/bib/bby121. Brief Bioinform. 2020. PMID: 30590436 Free PMC article. Review.
Cited by
-
Genome-wide unbalanced expression bias and expression level dominance toward Brassica oleracea in artificially synthesized intergeneric hybrids of Raphanobrassica.Hortic Res. 2021 Dec 1;8(1):246. doi: 10.1038/s41438-021-00672-2. Hortic Res. 2021. PMID: 34848691 Free PMC article.
-
Effects of Allopolyploidization and Homoeologous Chromosomal Segment Exchange on Homoeolog Expression in a Synthetic Allotetraploid Wheat under Variable Environmental Conditions.Plants (Basel). 2023 Aug 30;12(17):3111. doi: 10.3390/plants12173111. Plants (Basel). 2023. PMID: 37687357 Free PMC article.
-
Evolutionary divergence of subgenomes in common carp provides insights into speciation and allopolyploid success.Fundam Res. 2023 Aug 1;4(3):589-602. doi: 10.1016/j.fmre.2023.06.011. eCollection 2024 May. Fundam Res. 2023. PMID: 38933191 Free PMC article.
-
Evolution and Expression of the Expansin Genes in Emmer Wheat.Int J Mol Sci. 2023 Sep 15;24(18):14120. doi: 10.3390/ijms241814120. Int J Mol Sci. 2023. PMID: 37762423 Free PMC article.
-
Genomic and Meiotic Changes Accompanying Polyploidization.Plants (Basel). 2022 Jan 3;11(1):125. doi: 10.3390/plants11010125. Plants (Basel). 2022. PMID: 35009128 Free PMC article. Review.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

