Population Dynamics in Italian Canids between the Late Pleistocene and Bronze Age
- PMID: 33256122
- PMCID: PMC7761486
- DOI: 10.3390/genes11121409
Population Dynamics in Italian Canids between the Late Pleistocene and Bronze Age
Abstract
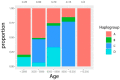
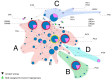
Dog domestication is still largely unresolved due to time-gaps in the sampling of regions. Ancient Italian canids are particularly understudied, currently represented by only a few specimens. In the present study, we sampled 27 canid remains from Northern Italy dated between the Late Pleistocene and Bronze Age to assess their genetic variability, and thus add context to dog domestication dynamics. They were targeted at four DNA fragments of the hypervariable region 1 of mitochondrial DNA. A total of 11 samples had good DNA preservation and were used for phylogenetic analyses. The dog samples were assigned to dog haplogroups A, C and D, and a Late Pleistocene wolf was set into wolf haplogroup 2. We present our data in the landscape of ancient and modern dog genetic variability, with a particular focus on the ancient Italian samples published thus far. Our results suggest there is high genetic variability within ancient Italian canids, where close relationships were evident between both a ~24,700 years old Italian canid, and Iberian and Bulgarian ancient dogs. These findings emphasize that disentangling dog domestication dynamics benefits from the analysis of specimens from Southern European regions.
Keywords: Italy; ancient DNA; archaeology; dogs; domestication; mitochondrial DNA; population genetics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Morey D.F., Jeger R. From wolf to dog: Late Pleistocene ecological dynamics, altered trophic strategies, and shifting human perceptions. Hist. Biol. 2017;29:895–903. doi: 10.1080/08912963.2016.1262854. - DOI
-
- Larson G., Karlsson E.K., Perri A., Webster M.T., Ho S.Y., Peters J., Stahl P.W., Piper P.J., Lingaas F., Fredholm M. Rethinking dog domestication by integrating genetics, archeology, and biogeography. Proc. Natl. Acad. Sci. USA. 2012;109:8878–8883. doi: 10.1073/pnas.1203005109. - DOI - PMC - PubMed
-
- Lallensack R. Ancient Genomes Heat up Dog Domestication Debate: Nature News & Comment. [(accessed on 5 February 2019)]; Available online: https://www.nature.com/news/ancient-genomes-heat-up-dog-domestication-de....
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

