Sequencing and analysis of gerbera daisy leaf transcriptomes reveal disease resistance and susceptibility genes differentially expressed and associated with powdery mildew resistance
- PMID: 33256589
- PMCID: PMC7706040
- DOI: 10.1186/s12870-020-02742-4
Sequencing and analysis of gerbera daisy leaf transcriptomes reveal disease resistance and susceptibility genes differentially expressed and associated with powdery mildew resistance
Abstract
Background: RNA sequencing has been widely used to profile genome-wide gene expression and identify candidate genes controlling disease resistance and other important traits in plants. Gerbera daisy is one of the most important flowers in the global floricultural trade, and powdery mildew (PM) is the most important disease of gerbera. Genetic improvement of gerbera PM resistance has become a crucial goal in gerbera breeding. A better understanding of the genetic control of gerbera resistance to PM can expedite the development of PM-resistant cultivars.
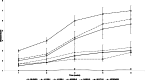
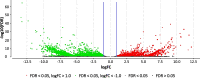
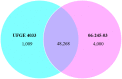
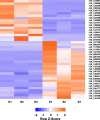
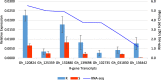
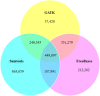
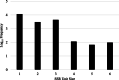
Results: The objectives of this study were to identify gerbera genotypes with contrasting phenotypes in PM resistance and sequence and analyze their leaf transcriptomes to identify disease resistance and susceptibility genes differentially expressed and associated with PM resistance. An additional objective was to identify SNPs and SSRs for use in future genetic studies. We identified two gerbera genotypes, UFGE 4033 and 06-245-03, that were resistant and susceptible to PM, respectively. De novo assembly of their leaf transcriptomes using four complementary pipelines resulted in 145,348 transcripts with a N50 of 1124 bp, of which 67,312 transcripts contained open reading frames and 48,268 were expressed in both genotypes. A total of 494 transcripts were likely involved in disease resistance, and 17 and 24 transcripts were up- and down-regulated, respectively, in UFGE 4033 compared to 06-245-03. These gerbera disease resistance transcripts were most similar to the NBS-LRR class of plant resistance genes conferring resistance to various pathogens in plants. Four disease susceptibility transcripts (MLO-like) were expressed only or highly expressed in 06-245-03, offering excellent candidate targets for gene editing for PM resistance in gerbera. A total of 449,897 SNPs and 19,393 SSRs were revealed in the gerbera transcriptomes, which can be a valuable resource for developing new molecular markers.
Conclusion: This study represents the first transcriptomic analysis of gerbera PM resistance, a highly important yet complex trait in a globally important floral crop. The differentially expressed disease resistance and susceptibility transcripts identified provide excellent targets for development of molecular markers and genetic maps, cloning of disease resistance genes, or targeted mutagenesis of disease susceptibility genes for PM resistance in gerbera.
Keywords: Differentially expressed genes; Disease resistance; Gerbera; Powdery mildew resistance; R-gene; Simple sequence repeats; Single nucleotide polymorphisms; Susceptibility gene.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Similar articles
-
Comparative Analysis of Impatiens Leaf Transcriptomes Reveal Candidate Genes for Resistance to Downy Mildew Caused by Plasmopara obducens.Int J Mol Sci. 2018 Jul 15;19(7):2057. doi: 10.3390/ijms19072057. Int J Mol Sci. 2018. PMID: 30011952 Free PMC article.
-
Construction of a genome-wide genetic linkage map and identification of quantitative trait loci for powdery mildew resistance in Gerbera daisy.Front Plant Sci. 2023 Jan 6;13:1072717. doi: 10.3389/fpls.2022.1072717. eCollection 2022. Front Plant Sci. 2023. PMID: 36684731 Free PMC article.
-
QTL-seq analysis of powdery mildew resistance in a Korean cucumber inbred line.Theor Appl Genet. 2021 Feb;134(2):435-451. doi: 10.1007/s00122-020-03705-x. Epub 2020 Oct 18. Theor Appl Genet. 2021. PMID: 33070226
-
Gene-Based Resistance to Erysiphe Species Causing Powdery Mildew Disease in Peas (Pisum sativum L.).Genes (Basel). 2022 Feb 8;13(2):316. doi: 10.3390/genes13020316. Genes (Basel). 2022. PMID: 35205360 Free PMC article. Review.
-
Research Advances in Genetic Mechanisms of Major Cucumber Diseases Resistance.Front Plant Sci. 2022 May 19;13:862486. doi: 10.3389/fpls.2022.862486. eCollection 2022. Front Plant Sci. 2022. PMID: 35665153 Free PMC article. Review.
Cited by
-
Cytological and transcriptomic analysis to unveil the mechanism of web blotch resistance in Peanut.BMC Plant Biol. 2023 Oct 26;23(1):518. doi: 10.1186/s12870-023-04545-9. BMC Plant Biol. 2023. PMID: 37884908 Free PMC article.
-
Polyketide Derivatives in the Resistance of Gerbera hybrida to Powdery Mildew.Front Plant Sci. 2022 Jan 6;12:790907. doi: 10.3389/fpls.2021.790907. eCollection 2021. Front Plant Sci. 2022. PMID: 35069647 Free PMC article.
-
Comparative transcriptome analysis of resistant and susceptible Kentucky bluegrass varieties in response to powdery mildew infection.BMC Plant Biol. 2022 Nov 2;22(1):509. doi: 10.1186/s12870-022-03883-4. BMC Plant Biol. 2022. PMID: 36319971 Free PMC article.
-
A dual transcriptome analysis reveals accession-specific resistance responses in Lathyrus sativus against Erysiphe pisi.Front Plant Sci. 2025 Mar 5;16:1542926. doi: 10.3389/fpls.2025.1542926. eCollection 2025. Front Plant Sci. 2025. PMID: 40110352 Free PMC article.
-
Genome-wide association study of Verticillium longisporum resistance in Brassica genotypes.Front Plant Sci. 2024 Aug 27;15:1436982. doi: 10.3389/fpls.2024.1436982. eCollection 2024. Front Plant Sci. 2024. PMID: 39258297 Free PMC article.
References
-
- Lynch RI. Gerbera, with a coloured plate of the new hybrids. Flora Sylva. 1905;3:206–208.
-
- FloraHolland. Facts and figures 2014. https://www.royalfloraholland.com/media/3949227/Kengetallen-2014-Engels.pdf. 2014. [Accessed December 12, 2015].
-
- United States Department of Agriculture (USDA). Floriculture Crops 2018 Summary. https://www.nass.usda.gov/Publications/Todays_Reports/reports/floran19.pdf. 2019. [Accessed November 6, 2019].
-
- GPN Magazine. https://gpnmag.com/article/new-standards-in-gerberas/. 2018. [Accessed May 15, 2020].
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

