Highly diversified core promoters in the human genome and their effects on gene expression and disease predisposition
- PMID: 33256598
- PMCID: PMC7706239
- DOI: 10.1186/s12864-020-07222-5
Highly diversified core promoters in the human genome and their effects on gene expression and disease predisposition
Abstract
Background: Core promoter controls transcription initiation. However, little is known for core promoter diversity in the human genome and its relationship with diseases. We hypothesized that as a functional important component in the genome, the core promoter in the human genome could be under evolutionary selection, as reflected by its highly diversification in order to adjust gene expression for better adaptation to the different environment.
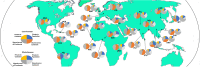
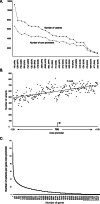
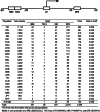
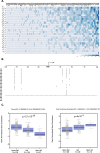
Results: Applying the "Exome-based Variant Detection in Core-promoters" method, we analyzed human core-promoter diversity by using the 2682 exome data sets of 25 worldwide human populations sequenced by the 1000 Genome Project. Collectively, we identified 31,996 variants in the core promoter region (- 100 to + 100) of 12,509 human genes ( https://dbhcpd.fhs.um.edu.mo ). Analyzing the rich variation data identified highly ethnic-specific patterns of core promoter variation between different ethnic populations, the genes with highly variable core promoters, the motifs affected by the variants, and their involved functional pathways. eQTL test revealed that 12% of core promoter variants can significantly alter gene expression level. Comparison with GWAS data we located 163 variants as the GWAS identified traits associated with multiple diseases, half of these variants can alter gene expression.
Conclusion: Data from our study reals the highly diversified nature of core promoter in the human genome, and highlights that core promoter variation could play important roles not only in gene expression regulation but also in disease predisposition.
Keywords: 1000 genomes; Core promoter; Exome; GWAS; Variation; eQTL.
Conflict of interest statement
All authors declare no competing interests.
Figures
Similar articles
-
Exome-based Variant Detection in Core Promoters.Sci Rep. 2016 Jul 28;6:30716. doi: 10.1038/srep30716. Sci Rep. 2016. PMID: 27464681 Free PMC article.
-
Natural selection in a population of Drosophila melanogaster explained by changes in gene expression caused by sequence variation in core promoter regions.BMC Evol Biol. 2016 Feb 9;16:35. doi: 10.1186/s12862-016-0606-3. BMC Evol Biol. 2016. PMID: 26860869 Free PMC article.
-
Prevalence of the initiator over the TATA box in human and yeast genes and identification of DNA motifs enriched in human TATA-less core promoters.Gene. 2007 Mar 1;389(1):52-65. doi: 10.1016/j.gene.2006.09.029. Epub 2006 Oct 10. Gene. 2007. PMID: 17123746 Free PMC article.
-
Variation in the Untranslated Genome and Susceptibility to Infections.Front Immunol. 2018 Sep 7;9:2046. doi: 10.3389/fimmu.2018.02046. eCollection 2018. Front Immunol. 2018. PMID: 30245696 Free PMC article. Review.
-
The human CYP19 (aromatase P450) gene: update on physiologic roles and genomic organization of promoters.J Steroid Biochem Mol Biol. 2003 Sep;86(3-5):219-24. doi: 10.1016/s0960-0760(03)00359-5. J Steroid Biochem Mol Biol. 2003. PMID: 14623514 Review.
Cited by
-
Core promoter in TNBC is highly mutated with rich ethnic signature.Brief Funct Genomics. 2023 Jan 20;22(1):9-19. doi: 10.1093/bfgp/elac035. Brief Funct Genomics. 2023. PMID: 36307127 Free PMC article.
-
Core promoter mutation contributes to abnormal gene expression in bladder cancer.BMC Cancer. 2022 Jan 15;22(1):68. doi: 10.1186/s12885-022-09178-z. BMC Cancer. 2022. PMID: 35033028 Free PMC article.
-
Etiological roles of core promoter variation in triple-negative breast cancer.Genes Dis. 2022 Feb 24;10(1):228-238. doi: 10.1016/j.gendis.2022.01.003. eCollection 2023 Jan. Genes Dis. 2022. PMID: 37013029 Free PMC article.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

