Isabl Platform, a digital biobank for processing multimodal patient data
- PMID: 33256603
- PMCID: PMC7708092
- DOI: 10.1186/s12859-020-03879-7
Isabl Platform, a digital biobank for processing multimodal patient data
Abstract
Background: The widespread adoption of high throughput technologies has democratized data generation. However, data processing in accordance with best practices remains challenging and the data capital often becomes siloed. This presents an opportunity to consolidate data assets into digital biobanks-ecosystems of readily accessible, structured, and annotated datasets that can be dynamically queried and analysed.
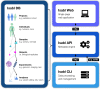
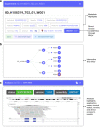
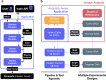
Results: We present Isabl, a customizable plug-and-play platform for the processing of multimodal patient-centric data. Isabl's architecture consists of a relational database (Isabl DB), a command line client (Isabl CLI), a RESTful API (Isabl API) and a frontend web application (Isabl Web). Isabl supports automated deployment of user-validated pipelines across the entire data capital. A full audit trail is maintained to secure data provenance, governance and ensuring reproducibility of findings.
Conclusions: As a digital biobank, Isabl supports continuous data utilization and automated meta analyses at scale, and serves as a catalyst for research innovation, new discoveries, and clinical translation.
Keywords: Analysis information management system; Data processing; Genomics; Image processing; Multimodal data; Next generation sequencing; Software engineering.
Conflict of interest statement
JSM, EP, and AK are founders of Isabl a whole genome analytics company.
Figures

References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

