Differential gene expression analysis reveals pathways important in early post-traumatic osteoarthritis in an equine model
- PMID: 33256611
- PMCID: PMC7708211
- DOI: 10.1186/s12864-020-07228-z
Differential gene expression analysis reveals pathways important in early post-traumatic osteoarthritis in an equine model
Abstract
Background: Post-traumatic osteoarthritis (PTOA) is a common and significant problem in equine athletes. It is a disease of the entire joint, with the synovium thought to be a key player in disease onset and progression due to its role in inflammation. The development of effective tools for early diagnosis and treatment of PTOA remains an elusive goal. Altered gene expression represents the earliest discernable disease-related change, and can provide valuable information about disease pathogenesis and identify potential therapeutic targets. However, there is limited work examining global gene expression changes in early disease. In this study, we quantified gene expression changes in the synovium of osteoarthritis-affected joints using an equine metacarpophalangeal joint (MCPJ) chip model of early PTOA. Synovial samples were collected arthroscopically from the MCPJ of 11 adult horses before (preOA) and after (OA) surgical induction of osteoarthritis and from sham-operated joints. After sequencing synovial RNA, Salmon was used to quasi-map reads and quantify transcript abundances. Differential expression analysis with the limma-trend method used a fold-change cutoff of log2(1.1). Functional annotation was performed with PANTHER at FDR < 0.05. Pathway and network analyses were performed in Reactome and STRING, respectively.
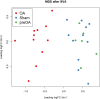
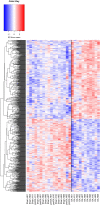
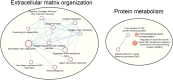
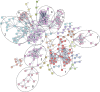
Results: RNA was sequenced from 28 samples (6 preOA, 11 OA, 11 sham). "Sham" and "preOA" were not different and were grouped. Three hundred ninety-seven genes were upregulated and 365 downregulated in OA synovium compared to unaffected. Gene ontology (GO) terms related to extracellular matrix (ECM) organization, angiogenesis, and cell signaling were overrepresented. There were 17 enriched pathways, involved in ECM turnover, protein metabolism, and growth factor signaling. Network analysis revealed clusters of differentially expressed genes involved in ECM organization, endothelial regulation, and cellular metabolism.
Conclusions: Enriched pathways and overrepresented GO terms reflected a state of high metabolic activity and tissue turnover in OA-affected tissue, suggesting that the synovium may retain the capacity to support healing and homeostasis in early disease. Limitations of this study include small sample size and capture of one point post-injury. Differentially expressed genes within key pathways may represent potential diagnostic markers or therapeutic targets for PTOA. Mechanistic validation of these findings is an important next step.
Keywords: Animal model; Degenerative joint disease; Metacarpophalangeal joint; Osteochondral fragment; RNAseq; Synovium.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Similar articles
-
Transient anabolic effects of synovium in early post-traumatic osteoarthritis: a novel ex vivo joint tissue co-culture system for investigating synovium-chondrocyte interactions.Osteoarthritis Cartilage. 2021 Jul;29(7):1060-1070. doi: 10.1016/j.joca.2021.03.010. Epub 2021 Mar 21. Osteoarthritis Cartilage. 2021. PMID: 33757858
-
RNA sequencing reveals the circular RNA expression profiles of osteoarthritic synovium.J Cell Biochem. 2019 Oct;120(10):18031-18040. doi: 10.1002/jcb.29106. Epub 2019 Jun 12. J Cell Biochem. 2019. PMID: 31190410
-
Laser Capture Microscopy RNA Sequencing for Topological Mapping of Synovial Pathology During Rheumatoid Arthritis.Arthritis Rheumatol. 2024 Aug;76(8):1243-1251. doi: 10.1002/art.42853. Epub 2024 May 21. Arthritis Rheumatol. 2024. PMID: 38556917 Free PMC article.
-
Inflammation in joint injury and post-traumatic osteoarthritis.Osteoarthritis Cartilage. 2015 Nov;23(11):1825-34. doi: 10.1016/j.joca.2015.08.015. Osteoarthritis Cartilage. 2015. PMID: 26521728 Free PMC article. Review.
-
Using mouse models to investigate the pathophysiology, treatment, and prevention of post-traumatic osteoarthritis.J Orthop Res. 2017 Mar;35(3):424-439. doi: 10.1002/jor.23343. Epub 2016 Jun 28. J Orthop Res. 2017. PMID: 27312470 Review.
Cited by
-
Modeling early changes associated with cartilage trauma using human-cell-laden hydrogel cartilage models.Stem Cell Res Ther. 2022 Aug 4;13(1):400. doi: 10.1186/s13287-022-03022-8. Stem Cell Res Ther. 2022. PMID: 35927702 Free PMC article.
-
Matrikine stimulation of equine synovial fibroblasts and chondrocytes results in an in vitro osteoarthritis phenotype.J Orthop Res. 2025 Feb;43(2):292-303. doi: 10.1002/jor.26004. Epub 2024 Nov 1. J Orthop Res. 2025. PMID: 39486895 Free PMC article.
-
The intersection of aging and estrogen in osteoarthritis.NPJ Womens Health. 2025;3(1):15. doi: 10.1038/s44294-025-00063-1. Epub 2025 Feb 25. NPJ Womens Health. 2025. PMID: 40017990 Free PMC article. Review.
-
Non-targeted metabolomic study in plasma in rats with post-traumatic osteoarthritis model.PLoS One. 2025 Mar 12;20(3):e0315708. doi: 10.1371/journal.pone.0315708. eCollection 2025. PLoS One. 2025. PMID: 40073326 Free PMC article.
-
RNAseq of Osteoarthritic Synovial Tissues: Systematic Literary Review.Front Aging. 2022 May 25;3:836791. doi: 10.3389/fragi.2022.836791. eCollection 2022. Front Aging. 2022. PMID: 35821799 Free PMC article.
References
-
- Martel-Pelletier J, Pelletier JP. Is osteoarthritis a disease involving only cartilage or other articular tissues? Eklem Hastalik Cerrahisi. 2010;21(1):2–14. - PubMed
-
- Neundorf RH, Lowerison MB, Cruz AM, Thomason JJ, McEwen BJ, Hurtig MB. Determination of the prevalence and severity of metacarpophalangeal joint osteoarthritis in thoroughbred racehorses via quantitative macroscopic evaluation. Am J Vet Res. 2010;71(11):1284–1293. doi: 10.2460/ajvr.71.11.1284. - DOI - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

