Epigenetic regulation of inflammation in periodontitis: cellular mechanisms and therapeutic potential
- PMID: 33256844
- PMCID: PMC7706209
- DOI: 10.1186/s13148-020-00982-7
Epigenetic regulation of inflammation in periodontitis: cellular mechanisms and therapeutic potential
Abstract
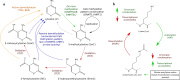
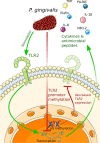
Epigenetic mechanisms, namely DNA and histone modifications, are critical regulators of immunity and inflammation which have emerged as potential targets for immunomodulating therapies. The prevalence and significant morbidity of periodontitis, in combination with accumulating evidence that genetic, environmental and lifestyle factors cannot fully explain the susceptibility of individuals to disease development, have driven interest in epigenetic regulation as an important factor in periodontitis pathogenesis. Aberrant promoter methylation profiles of genes involved in inflammatory activation, including TLR2, PTGS2, IFNG, IL6, IL8, and TNF, have been observed in the gingival tissue, peripheral blood or buccal mucosa from patients with periodontitis, correlating with changes in expression and disease severity. The expression of enzymes that regulate histone acetylation, in particular histone deacetylases (HDACs), is also dysregulated in periodontitis-affected gingival tissue. Infection of gingival epithelial cells, gingival fibroblasts and periodontal ligament cells with the oral pathogens Porphyromonas gingivalis or Treponema denticola induces alterations in expression and activity of chromatin-modifying enzymes, as well as site-specific and global changes in DNA methylation profiles and in histone acetylation and methylation marks. These epigenetic changes are associated with excessive production of inflammatory cytokines, chemokines, and matrix-degrading enzymes that can be suppressed by small molecule inhibitors of HDACs (HDACi) or DNA methyltransferases. HDACi and inhibitors of bromodomain-containing BET proteins ameliorate inflammation, osteoclastogenesis, and alveolar bone resorption in animal models of periodontitis, suggesting their clinical potential as host modulation therapeutic agents. However, broader application of epigenomic methods will be required to create a comprehensive map of epigenetic changes in periodontitis. The integration of functional studies with global analyses of the epigenetic landscape will provide critical information on the therapeutic and diagnostic potential of epigenetics in periodontal disease.
Keywords: Bromodomain; DNA methylation; Epigenetics; Histone deacetylase; Inflammation; Periodontitis; Porphyromonas gingivalis.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Similar articles
-
BET Bromodomain Inhibitors Suppress Inflammatory Activation of Gingival Fibroblasts and Epithelial Cells From Periodontitis Patients.Front Immunol. 2019 Apr 30;10:933. doi: 10.3389/fimmu.2019.00933. eCollection 2019. Front Immunol. 2019. PMID: 31114581 Free PMC article.
-
HDAC3 Regulates Gingival Fibroblast Inflammatory Responses in Periodontitis.J Dent Res. 2020 Jan;99(1):98-106. doi: 10.1177/0022034519885088. Epub 2019 Nov 6. J Dent Res. 2020. PMID: 31693860 Free PMC article.
-
TLR2 promoter hypermethylation creates innate immune dysbiosis.J Dent Res. 2015 Jan;94(1):183-91. doi: 10.1177/0022034514557545. Epub 2014 Nov 11. J Dent Res. 2015. PMID: 25389002 Free PMC article.
-
The Relevance of DNA Methylation and Histone Modification in Periodontitis: A Scoping Review.Cells. 2022 Oct 13;11(20):3211. doi: 10.3390/cells11203211. Cells. 2022. PMID: 36291079 Free PMC article.
-
[Pathogenic potential of Porphyromonas gingivalis, Treponema denticola and Tannerella forsythia, the red bacterial complex associated with periodontitis].Pathol Biol (Paris). 2007 Apr-May;55(3-4):154-62. doi: 10.1016/j.patbio.2006.07.045. Epub 2006 Oct 17. Pathol Biol (Paris). 2007. PMID: 17049750 Review. French.
Cited by
-
Palmitate-Triggered COX2/PGE2-Related Hyperinflammation in Dual-Stressed PdL Fibroblasts Is Mediated by Repressive H3K27 Trimethylation.Cells. 2022 Mar 10;11(6):955. doi: 10.3390/cells11060955. Cells. 2022. PMID: 35326406 Free PMC article.
-
Association between Maternal Periodontitis and Development of Systematic Diseases in Offspring.Int J Mol Sci. 2022 Feb 24;23(5):2473. doi: 10.3390/ijms23052473. Int J Mol Sci. 2022. PMID: 35269617 Free PMC article. Review.
-
The Receptor AT1 Appears to Be Important for the Maintenance of Bone Mass and AT2 Receptor Function in Periodontal Bone Loss Appears to Be Regulated by AT1 Receptor.Int J Mol Sci. 2021 Nov 27;22(23):12849. doi: 10.3390/ijms222312849. Int J Mol Sci. 2021. PMID: 34884653 Free PMC article.
-
The Emerging Regulatory Role of Circular RNAs in Periodontal Tissues and Cells.Int J Mol Sci. 2021 Apr 28;22(9):4636. doi: 10.3390/ijms22094636. Int J Mol Sci. 2021. PMID: 33924932 Free PMC article. Review.
-
DNMT3B (rs2424913) polymorphism is associated with systemic lupus erythematosus alone and with co-existing periodontitis in a Brazilian population.J Appl Oral Sci. 2022 Apr 29;30:e20210567. doi: 10.1590/1678-7757-2021-0567. eCollection 2022. J Appl Oral Sci. 2022. PMID: 35507987 Free PMC article.
References
-
- Preshaw PM. Host modulation therapy with anti-inflammatory agents. Periodontology. 2000;2018(76):131–149. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

