Microbial Lipid A Remodeling Controls Cross-Presentation Efficiency and CD8 T Cell Priming by Modulating Dendritic Cell Function
- PMID: 33257533
- PMCID: PMC7822130
- DOI: 10.1128/IAI.00335-20
Microbial Lipid A Remodeling Controls Cross-Presentation Efficiency and CD8 T Cell Priming by Modulating Dendritic Cell Function
Abstract
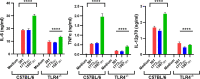
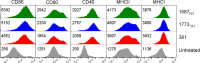
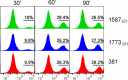
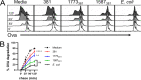
The majority of Gram-negative bacteria elicit a potent immune response via recognition of lipid A expressed on the outer bacterial membrane by the host immune receptor Toll-like receptor 4 (TLR4). However, some Gram-negative bacteria evade detection by TLR4 or alter the outcome of TLR4 signaling by modification of lipid A species. Although the role of lipid A modifications on host innate immunity has been examined in some detail, it is currently unclear how lipid A remodeling influences host adaptive immunity. One prototypic Gram-negative bacterium that modifies its lipid A structure is Porphyromonas gingivalis, an anaerobic pathobiont that colonizes the human periodontium and induces chronic low-grade inflammation that is associated with periodontal disease as well as a number of systemic inflammatory disorders. P. gingivalis produces dephosphorylated and deacylated lipid A structures displaying altered activities at TLR4. Here, we explored the functional role of P. gingivalis lipid A modifications on TLR4-dependent innate and adaptive immune responses in mouse bone marrow-derived dendritic cells (BMDCs). We discovered that lipid A 4'-phosphate removal is required for P. gingivalis to evade BMDC-dependent proinflammatory cytokine responses and markedly limits the bacterium's capacity to induce beta interferon (IFN-β) production. In addition, lipid A 4'-phosphatase activity prevents canonical bacterium-induced delay in antigen degradation, which leads to inefficient antigen cross-presentation and a failure to cross-prime CD8 T cells specific for a P. gingivalis-associated antigen. We propose that lipid A modifications produced by this bacterium alter host TLR4-dependent adaptive immunity to establish chronic infections associated with a number of systemic inflammatory disorders.
Keywords: Porphyromonas gingivalis; TLR4; antigen cross-presentation; bacterial pathogenesis; dendritic cells; inflammation; lipid A.
Copyright © 2021 American Society for Microbiology.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

