Essential omega-3 fatty acids tune microglial phagocytosis of synaptic elements in the mouse developing brain
- PMID: 33257673
- PMCID: PMC7704669
- DOI: 10.1038/s41467-020-19861-z
Essential omega-3 fatty acids tune microglial phagocytosis of synaptic elements in the mouse developing brain
Abstract
Omega-3 fatty acids (n-3 PUFAs) are essential for the functional maturation of the brain. Westernization of dietary habits in both developed and developing countries is accompanied by a progressive reduction in dietary intake of n-3 PUFAs. Low maternal intake of n-3 PUFAs has been linked to neurodevelopmental diseases in Humans. However, the n-3 PUFAs deficiency-mediated mechanisms affecting the development of the central nervous system are poorly understood. Active microglial engulfment of synapses regulates brain development. Impaired synaptic pruning is associated with several neurodevelopmental disorders. Here, we identify a molecular mechanism for detrimental effects of low maternal n-3 PUFA intake on hippocampal development in mice. Our results show that maternal dietary n-3 PUFA deficiency increases microglia-mediated phagocytosis of synaptic elements in the rodent developing hippocampus, partly through the activation of 12/15-lipoxygenase (LOX)/12-HETE signaling, altering neuronal morphology and affecting cognitive performance of the offspring. These findings provide a mechanistic insight into neurodevelopmental defects caused by maternal n-3 PUFAs dietary deficiency.
Conflict of interest statement
The authors declare no competing interests.
Figures
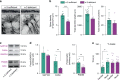
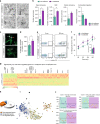

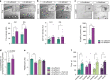
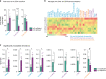

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

