A DNA-based vaccine protects against Crimean-Congo haemorrhagic fever virus disease in a Cynomolgus macaque model
- PMID: 33257849
- PMCID: PMC7854975
- DOI: 10.1038/s41564-020-00815-6
A DNA-based vaccine protects against Crimean-Congo haemorrhagic fever virus disease in a Cynomolgus macaque model
Abstract
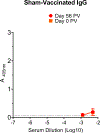
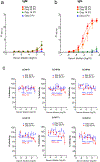
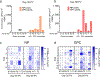
There is currently no specific prophylaxis or vaccine against Crimean-Congo haemorrhagic fever virus (CCHFV). Crimean-Congo haemorrhagic fever (CCHF) is a severe febrile illness transmitted by Hyalomma ticks in endemic areas, handling of infected livestock or care of infected patients. We report here the successful protection against CCHFV-mediated disease in a non-human primate disease model. Cynomolgus macaques were vaccinated with a DNA-based vaccine using in vivo electroporation-assisted delivery. The vaccine contained two plasmids encoding the glycoprotein precursor (GPC) and the nucleoprotein (NP) of CCHFV. Animals received three vaccinations and we recorded potent antibody and T cell responses after vaccination. While all sham-vaccinated animals developed viraemia, high tissue viral loads and CCHF-induced disease, the NP + GPC vaccinated animals were significantly protected. In conclusion, this is evidence of a vaccine that can protect against CCHFV-induced disease in a non-human primate model. This supports clinical development of the vaccine to protect groups at risk for contracting the infection.
Conflict of interest statement
Competing Interests
MS is a founder and chairman of the board of Svenska Vaccinfabriken Produktion AB.
Figures



Similar articles
-
Accelerated DNA vaccine regimen provides protection against Crimean-Congo hemorrhagic fever virus challenge in a macaque model.Mol Ther. 2023 Feb 1;31(2):387-397. doi: 10.1016/j.ymthe.2022.09.016. Epub 2022 Oct 3. Mol Ther. 2023. PMID: 36184852 Free PMC article.
-
Nucleoside-Modified mRNA Vaccines Protect IFNAR-/- Mice against Crimean-Congo Hemorrhagic Fever Virus Infection.J Virol. 2022 Feb 9;96(3):e0156821. doi: 10.1128/JVI.01568-21. Epub 2021 Nov 24. J Virol. 2022. PMID: 34817199 Free PMC article.
-
A DNA vaccine for Crimean-Congo hemorrhagic fever protects against disease and death in two lethal mouse models.PLoS Negl Trop Dis. 2017 Sep 18;11(9):e0005908. doi: 10.1371/journal.pntd.0005908. eCollection 2017 Sep. PLoS Negl Trop Dis. 2017. PMID: 28922426 Free PMC article.
-
Development of vaccines against Crimean-Congo haemorrhagic fever virus.Vaccine. 2017 Oct 20;35(44):6015-6023. doi: 10.1016/j.vaccine.2017.05.031. Epub 2017 Jul 4. Vaccine. 2017. PMID: 28687403 Free PMC article. Review.
-
Crimean-Congo haemorrhagic fever virus: Past, present and future insights for animal modelling and medical countermeasures.Zoonoses Public Health. 2018 Aug;65(5):465-480. doi: 10.1111/zph.12469. Epub 2018 Apr 20. Zoonoses Public Health. 2018. PMID: 29676526 Free PMC article. Review.
Cited by
-
Replicating RNA vaccination elicits an unexpected immune response that efficiently protects mice against lethal Crimean-Congo hemorrhagic fever virus challenge.EBioMedicine. 2022 Aug;82:104188. doi: 10.1016/j.ebiom.2022.104188. Epub 2022 Jul 27. EBioMedicine. 2022. PMID: 35907368 Free PMC article.
-
Replicon particle vaccination induces non-neutralizing anti-nucleoprotein antibody-mediated control of Crimean-Congo hemorrhagic fever virus.NPJ Vaccines. 2024 May 23;9(1):88. doi: 10.1038/s41541-024-00877-1. NPJ Vaccines. 2024. PMID: 38782933 Free PMC article.
-
The Integration of Human and Veterinary Studies for Better Understanding and Management of Crimean-Congo Haemorrhagic Fever.Front Immunol. 2021 Mar 18;12:629636. doi: 10.3389/fimmu.2021.629636. eCollection 2021. Front Immunol. 2021. PMID: 33815379 Free PMC article. Review.
-
Design and Analysis of Novel HEV Vaccine Variants and Evaluation of Two Selected Candidates in a Porcine Infection Model.Liver Int. 2025 Sep;45(9):e70246. doi: 10.1111/liv.70246. Liver Int. 2025. PMID: 40747926 Free PMC article.
-
GEM-PA-Based Subunit Vaccines of Crimean Congo Hemorrhagic Fever Induces Systemic Immune Responses in Mice.Viruses. 2022 Jul 28;14(8):1664. doi: 10.3390/v14081664. Viruses. 2022. PMID: 36016285 Free PMC article.
References
-
- Stuart MC, Kouimtzi M & Hill SR WHO Model Formulary 2008. (World Health Organization, 2008).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

