Double strand breaks (DSBs) as indicators of genomic instability in PATRR-mediated translocations
- PMID: 33258468
- PMCID: PMC7906754
- DOI: 10.1093/hmg/ddaa251
Double strand breaks (DSBs) as indicators of genomic instability in PATRR-mediated translocations
Abstract
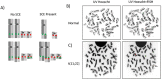
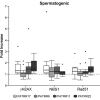
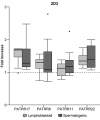
Genomic instability contributes to a variety of potentially damaging conditions, including DNA-based rearrangements. Breakage in the form of double strand breaks (DSBs) increases the likelihood of DNA damage, mutations and translocations. Certain human DNA regions are known to be involved in recurrent translocations, such as the palindrome-mediated rearrangements that have been identified at the breakpoints of several recurrent constitutional translocations: t(11;22)(q23;q11), t(17;22)(q11;q11) and t(8;22) (q24;q11). These breakpoints occur at the center of palindromic AT-rich repeats (PATRRs), which suggests that the structure of the DNA may play a contributory role, potentially through the formation of secondary cruciform structures. The current study analyzed the DSB propensity of these PATRR regions in both lymphoblastoid (mitotic) and spermatogenic cells (meiotic). Initial results found an increased association of sister chromatid exchanges (SCEs) at PATRR regions in experiments that used SCEs to assay DSBs, combining SCE staining with fluorescence in situ hybridization (FISH). Additional experiments used chromatin immunoprecipitation (ChIP) with antibodies for either markers of DSBs or proteins involved in DSB repair along with quantitative polymerase chain reaction to quantify the frequency of DSBs occurring at PATRR regions. The results indicate an increased rate of DSBs at PATRR regions. Additional ChIP experiments with the cruciform binding 2D3 antibody indicate an increased rate of cruciform structures at PATRR regions in both mitotic and meiotic samples. Overall, these experiments demonstrate an elevated rate of DSBs at PATRR regions, an indication that the structure of PATRR containing DNA may lead to increased breakage in multiple cellular environments.
© The Author(s) 2020. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures
Similar articles
-
Palindrome-mediated chromosomal translocations in humans.DNA Repair (Amst). 2006 Sep 8;5(9-10):1136-45. doi: 10.1016/j.dnarep.2006.05.035. Epub 2006 Jul 10. DNA Repair (Amst). 2006. PMID: 16829213 Free PMC article. Review.
-
Chromosomal translocations and palindromic AT-rich repeats.Curr Opin Genet Dev. 2012 Jun;22(3):221-8. doi: 10.1016/j.gde.2012.02.004. Epub 2012 Mar 6. Curr Opin Genet Dev. 2012. PMID: 22402448 Free PMC article. Review.
-
Long AT-rich palindromes and the constitutional t(11;22) breakpoint.Hum Mol Genet. 2001 Nov 1;10(23):2605-17. doi: 10.1093/hmg/10.23.2605. Hum Mol Genet. 2001. PMID: 11726547
-
Cruciform DNA structure underlies the etiology for palindrome-mediated human chromosomal translocations.J Biol Chem. 2004 Aug 20;279(34):35377-83. doi: 10.1074/jbc.M400354200. Epub 2004 Jun 20. J Biol Chem. 2004. PMID: 15208332 Free PMC article.
-
Chromosomal translocations mediated by palindromic DNA.Cell Cycle. 2006 Jun;5(12):1297-303. doi: 10.4161/cc.5.12.2809. Epub 2006 Jun 15. Cell Cycle. 2006. PMID: 16760666 Review.
Cited by
-
Interaction of Proteins with Inverted Repeats and Cruciform Structures in Nucleic Acids.Int J Mol Sci. 2022 May 31;23(11):6171. doi: 10.3390/ijms23116171. Int J Mol Sci. 2022. PMID: 35682854 Free PMC article. Review.
-
A Novel Non-Allelic Homologous Recombination Event in a Parent with an 11;22 Reciprocal Translocation Leading to 22q11.2 Deletion Syndrome.Genes (Basel). 2022 Sep 17;13(9):1668. doi: 10.3390/genes13091668. Genes (Basel). 2022. PMID: 36140835 Free PMC article.
-
Combining long-read DNA and RNA sequencing to enhance molecular understanding of structural variations leading to copy gains.Comput Struct Biotechnol J. 2025 Apr 24;27:1732-1740. doi: 10.1016/j.csbj.2025.04.031. eCollection 2025. Comput Struct Biotechnol J. 2025. PMID: 40421160 Free PMC article.
-
Downstream transcription promotes human recurrent CNV associated AT-rich sequence mediated genome rearrangements in yeast.iScience. 2024 Nov 30;27(12):111508. doi: 10.1016/j.isci.2024.111508. eCollection 2024 Dec 20. iScience. 2024. PMID: 39758996 Free PMC article.
-
Palindromes in DNA-A Risk for Genome Stability and Implications in Cancer.Int J Mol Sci. 2021 Mar 11;22(6):2840. doi: 10.3390/ijms22062840. Int J Mol Sci. 2021. PMID: 33799581 Free PMC article. Review.
References
-
- Zackai, E.H. and Emanuel, B.S. (1980) Site-specific reciprocal translocation, t(11;22) (q23;q11), in several unrelated families with 3:1 meiotic disjunction. Am. J. Med. Genet., 7, 507–521. - PubMed
-
- Fraccaro, M., Lindsten, J., Ford, C.E. and Iselius, L. (1980) The 11q;22q translocation: a European collaborative analysis of 43 cases. Hum. Genet., 56, 21–51. - PubMed

