Protein phosphatase NtPP2C2b and MAP kinase NtMPK4 act in concert to modulate nicotine biosynthesis
- PMID: 33258946
- PMCID: PMC7921305
- DOI: 10.1093/jxb/eraa568
Protein phosphatase NtPP2C2b and MAP kinase NtMPK4 act in concert to modulate nicotine biosynthesis
Abstract
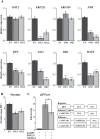
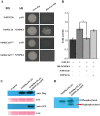
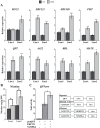
Protein phosphatases (PPs) and protein kinases (PKs) regulate numerous developmental, defense, and phytohormone signaling processes in plants. However, the underlying regulatory mechanism governing biosynthesis of specialized metabolites, such as alkaloids, by the combined effects of PPs and PKs, is insufficiently understood. Here, we report the characterization of a group B protein phosphatase type 2C, NtPP2C2b, that likely acts upstream of the NICOTINE2 locus APETALA 2/Ethylene Response Factors (AP2/ERFs), to regulate nicotine biosynthesis in tobacco. Similar to the nicotine pathway genes, NtPP2C2b is highly expressed in roots and induced by jasmonic acid (JA). Overexpression of NtPP2C2b in transgenic hairy roots or stable transgenic tobacco plants repressed nicotine pathway gene expression and reduced nicotine accumulation. Additionally, transient overexpression of NtPP2C2b, together with the NtERF221, repressed transactivation of the quinolinate phosphoribosyltransferase promoter in tobacco cells. We further demonstrate that the JA-responsive tobacco mitogen-activated protein kinase (MAPK) 4 interacts with NtPP2C2b in yeast and plant cells. Conditional overexpression of NtMPK4 in tobacco hairy roots up-regulated nicotine pathway gene expression and increased nicotine accumulation. Our findings suggest that a previously uncharacterized PP-PK module acts to modulate alkaloid biosynthesis, highlighting the importance of post-translational control in the biosynthesis of specialized plant metabolites.
Keywords: Alkaloid biosynthesis; MAP kinase; gene regulation; hairy roots; nicotine; protein phosphatase 2C; secondary metabolism; tobacco.
© The Author(s) 2020. Published by Oxford University Press on behalf of the Society for Experimental Biology.
Figures



Similar articles
-
APETALA2/ETHYLENE RESPONSE FACTOR and basic helix-loop-helix tobacco transcription factors cooperatively mediate jasmonate-elicited nicotine biosynthesis.Plant J. 2011 Jun;66(6):1053-65. doi: 10.1111/j.1365-313X.2011.04566.x. Epub 2011 Apr 5. Plant J. 2011. PMID: 21418355
-
Tobacco MYC2 regulates jasmonate-inducible nicotine biosynthesis genes directly and by way of the NIC2-locus ERF genes.Plant Cell Physiol. 2011 Jun;52(6):1117-30. doi: 10.1093/pcp/pcr063. Epub 2011 May 16. Plant Cell Physiol. 2011. PMID: 21576194
-
Stress-induced expression of NICOTINE2-locus genes and their homologs encoding Ethylene Response Factor transcription factors in tobacco.Phytochemistry. 2015 May;113:41-9. doi: 10.1016/j.phytochem.2014.05.017. Epub 2014 Jun 16. Phytochemistry. 2015. PMID: 24947337
-
Molecular genetics of alkaloid biosynthesis in Nicotiana tabacum.Phytochemistry. 2013 Oct;94:10-27. doi: 10.1016/j.phytochem.2013.06.002. Epub 2013 Aug 15. Phytochemistry. 2013. PMID: 23953973 Review.
-
[Research progress of jasmonate-responsive transcription factors in regulating plant secondary metabolism].Zhongguo Zhong Yao Za Zhi. 2018 Mar;43(5):897-903. doi: 10.19540/j.cnki.cjcmm.20180109.001. Zhongguo Zhong Yao Za Zhi. 2018. PMID: 29676085 Review. Chinese.
Cited by
-
Interplay of transcription factors orchestrating the biosynthesis of plant alkaloids.3 Biotech. 2022 Oct;12(10):250. doi: 10.1007/s13205-022-03316-x. Epub 2022 Aug 29. 3 Biotech. 2022. PMID: 36051988 Free PMC article. Review.
-
Light regulation of the biosynthesis of phenolics, terpenoids, and alkaloids in plants.Commun Biol. 2023 Oct 18;6(1):1055. doi: 10.1038/s42003-023-05435-4. Commun Biol. 2023. PMID: 37853112 Free PMC article. Review.
-
ZmPP2C26 Alternative Splicing Variants Negatively Regulate Drought Tolerance in Maize.Front Plant Sci. 2022 Apr 8;13:851531. doi: 10.3389/fpls.2022.851531. eCollection 2022. Front Plant Sci. 2022. PMID: 35463404 Free PMC article.
-
Reprogramming plant specialized metabolism by manipulating protein kinases.aBIOTECH. 2021;2(3):226-239. doi: 10.1007/s42994-021-00053-2. Epub 2021 Jun 17. aBIOTECH. 2021. PMID: 34377580 Free PMC article. Review.
-
Genetic regulation and manipulation of nicotine biosynthesis in tobacco: strategies to eliminate addictive alkaloids.J Exp Bot. 2024 Mar 14;75(6):1741-1753. doi: 10.1093/jxb/erad341. J Exp Bot. 2024. PMID: 37647764 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

