A Two-Locus System with Strong Epistasis Underlies Rapid Parasite-Mediated Evolution of Host Resistance
- PMID: 33258959
- PMCID: PMC8042741
- DOI: 10.1093/molbev/msaa311
A Two-Locus System with Strong Epistasis Underlies Rapid Parasite-Mediated Evolution of Host Resistance
Erratum in
-
Erratum to: A Two-Locus System with Strong Epistasis Underlies Rapid Parasite-Mediated Evolution of Host Resistance.Mol Biol Evol. 2021 May 19;38(6):2674. doi: 10.1093/molbev/msab076. Mol Biol Evol. 2021. PMID: 33846744 Free PMC article. No abstract available.
Abstract
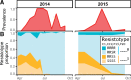
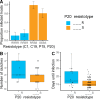
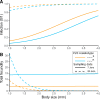
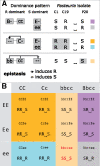
Parasites are a major evolutionary force, driving adaptive responses in host populations. Although the link between phenotypic response to parasite-mediated natural selection and the underlying genetic architecture often remains obscure, this link is crucial for understanding the evolution of resistance and predicting associated allele frequency changes in the population. To close this gap, we monitored the response to selection during epidemics of a virulent bacterial pathogen, Pasteuria ramosa, in a natural host population of Daphnia magna. Across two epidemics, we observed a strong increase in the proportion of resistant phenotypes as the epidemics progressed. Field and laboratory experiments confirmed that this increase in resistance was caused by selection from the local parasite. Using a genome-wide association study, we built a genetic model in which two genomic regions with dominance and epistasis control resistance polymorphism in the host. We verified this model by selfing host genotypes with different resistance phenotypes and scoring their F1 for segregation of resistance and associated genetic markers. Such epistatic effects with strong fitness consequences in host-parasite coevolution are believed to be crucial in the Red Queen model for the evolution of genetic recombination.
Keywords: Daphnia magna; Pasteuria ramosa; dominance; epistasis; genetic architecture; multilocus genetics; parasite-mediated selection; resistance; zooplankton.
© The Author(s) 2020. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures

References
-
- Andras JP, Ebert D.. 2013. A novel approach to parasite population genetics: experimental infection reveals geographic differentiation, recombination and host-mediated population structure in Pasteuria ramosa, a bacterial parasite of Daphnia. Mol Ecol. 22(4):972–986. - PubMed
-
- Auguie B. 2017. gridExtra: miscellaneous functions for “Grid” graphics. Available from: https://CRAN.R-project.org/package=gridExtra. Last accessed on December 2020.
-
- Auguie B. 2019. egg: extensions for “ggplot2”: custom geom, custom themes, plot alignment, labelled panels, symmetric scales, and fixed panel size. Available from: https://CRAN.R-project.org/package=egg. Last accessed on December 2020.
Publication types
MeSH terms
Supplementary concepts
Associated data
LinkOut - more resources
Full Text Sources
Miscellaneous

