Zinc Binds to RRM2 Peptide of TDP-43
- PMID: 33260324
- PMCID: PMC7730363
- DOI: 10.3390/ijms21239080
Zinc Binds to RRM2 Peptide of TDP-43
Abstract
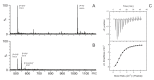
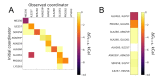
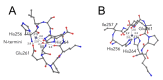
Transactive response DNA and RNA binding protein 43 kDa (TDP-43) is a highly conserved heterogeneous nuclear ribonucleoprotein (hnRNP), which is involved in several steps of protein production including transcription and splicing. Its aggregates are frequently observed in motor neurons from amyotrophic lateral sclerosis patients and in the most common variant of frontotemporal lobar degeneration. Recently it was shown that TDP-43 is able to bind Zn2+ by its RRM domain. In this work, we have investigated Zn2+ binding to a short peptide 256-264 from C-terminus of RRM2 domain using isothermal titration calorimetry, electrospray ionization mass spectrometry, QM/MM simulations, and NMR spectroscopy. We have found that this peptide is able to bind zinc ions with a Ka equal to 1.6 × 105 M-1. Our findings suggest the existence of a zinc binding site in the C-terminal region of RRM2 domain. Together with the existing structure of the RRM2 domain of TDP-43 we propose a model of its complex with Zn2+ which illustrates how zinc might regulate DNA/RNA binding.
Keywords: QM/MM; TDP-43; zinc.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Tsvetkov P.O., Roman A.Y., Baksheeva V.E., Nazipova A.A., Shevelyova M.P., Vladimirov V.I., Buyanova M.F., Zinchenko D.V., Zamyatnin A.A.J., Devred F., et al. Functional Status of Neuronal Calcium Sensor-1 Is Modulated by Zinc Binding. Front. Mol. Neurosci. 2018;11:459. doi: 10.3389/fnmol.2018.00459. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

