Assessment of Viral Targeted Sequence Capture Using Nanopore Sequencing Directly from Clinical Samples
- PMID: 33260903
- PMCID: PMC7759923
- DOI: 10.3390/v12121358
Assessment of Viral Targeted Sequence Capture Using Nanopore Sequencing Directly from Clinical Samples
Abstract
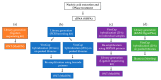
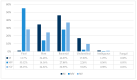
Shotgun metagenomic sequencing (SMg) enables the simultaneous detection and characterization of viruses in human, animal and environmental samples. However, lack of sensitivity still poses a challenge and may lead to poor detection and data acquisition for detailed analysis. To improve sensitivity, we assessed a broad scope targeted sequence capture (TSC) panel (ViroCap) in both human and animal samples. Moreover, we adjusted TSC for the Oxford Nanopore MinION and compared the performance to an SMg approach. TSC on the Illumina NextSeq served as the gold standard. Overall, TSC increased the viral read count significantly in challenging human samples, with the highest genome coverage achieved using the TSC on the MinION. TSC also improved the genome coverage and sequencing depth in clinically relevant viruses in the animal samples, such as influenza A virus. However, SMg was shown to be adequate for characterizing a highly diverse animal virome. TSC on the MinION was comparable to the NextSeq and can provide a valuable alternative, offering longer reads, portability and lower initial cost. Developing new viral enrichment approaches to detect and characterize significant human and animal viruses is essential for the One Health Initiative.
Keywords: next-generation sequencing; one health; porcine viruses; shotgun metagenomic sequencing; targeted sequence capture; viral metagenomics; virome.
Conflict of interest statement
John W. A. Rossen is employed by IDbyDNA. Silke Peter consults for IDbyDNA. This did not have any influence on the interpretation of reviewed data and conclusions drawn nor on the drafting of the manuscript, and no support was obtained from them. All other authors declare no conflict of interest.
Figures

References
-
- Meredith L.W., Hamilton W.L., Warne B., Houldcroft C.J., Hosmillo M., Jahun A., Curran M.D., Parmar S., Caller L., Caddy S.L. Rapid implementation of real-time SARS-CoV-2 sequencing to investigate healthcare-associated COVID-19 infections. MedRxiv. 2020;20 doi: 10.1016/S1473-3099(20)30562-4. - DOI - PMC - PubMed
-
- Couto N., Schuele L., Raangs E.C., Machado M.P., Mendes C.I., Jesus T.F., Chlebowicz M., Rosema S., Ramirez M., Carriço J.A. Critical steps in clinical shotgun metagenomics for the concomitant detection and typing of microbial pathogens. Sci. Rep. 2018;8:13767. doi: 10.1038/s41598-018-31873-w. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

