Cannabinoid Receptor Interacting Protein 1a (CRIP1a) in Health and Disease
- PMID: 33261012
- PMCID: PMC7761089
- DOI: 10.3390/biom10121609
Cannabinoid Receptor Interacting Protein 1a (CRIP1a) in Health and Disease
Abstract
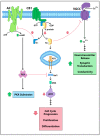
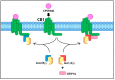
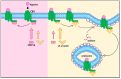
Endocannabinoid signaling depends upon the CB1 and CB2 cannabinoid receptors, their endogenous ligands anandamide and 2-arachidonoylglycerol, and intracellular proteins that mediate responses via the C-terminal and other intracellular receptor domains. The CB1 receptor regulates and is regulated by associated G proteins predominantly of the Gi/o subtypes, β-arrestins 1 and 2, and the cannabinoid receptor-interacting protein 1a (CRIP1a). Evidence for a physiological role for CRIP1a is emerging as data regarding the cellular localization and function of CRIP1a are generated. Here we summarize the neuronal distribution and role of CRIP1a in endocannabinoid signaling, as well as discuss investigations linking CRIP1a to development, vision and hearing sensory systems, hippocampus and seizure regulation, and psychiatric disorders including schizophrenia. We also examine the genetic and epigenetic association of CRIP1a within a variety of cancer subtypes. This review provides evidence upon which to base future investigations on the function of CRIP1a in health and disease.
Keywords: G protein-coupled receptors (GPCRs), hippocampus; cancer; embryonic development; endocannabinoids; epilepsy; retina; schizophrenia; seizures.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

