Antisense Transcription across Nucleotide Repeat Expansions in Neurodegenerative and Neuromuscular Diseases: Progress and Mysteries
- PMID: 33261024
- PMCID: PMC7760973
- DOI: 10.3390/genes11121418
Antisense Transcription across Nucleotide Repeat Expansions in Neurodegenerative and Neuromuscular Diseases: Progress and Mysteries
Abstract
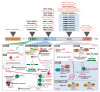
Unstable repeat expansions and insertions cause more than 30 neurodegenerative and neuromuscular diseases. Remarkably, bidirectional transcription of repeat expansions has been identified in at least 14 of these diseases. More remarkably, a growing number of studies has been showing that both sense and antisense repeat RNAs are able to dysregulate important cellular pathways, contributing together to the observed clinical phenotype. Notably, antisense repeat RNAs from spinocerebellar ataxia type 7, myotonic dystrophy type 1, Huntington's disease and frontotemporal dementia/amyotrophic lateral sclerosis associated genes have been implicated in transcriptional regulation of sense gene expression, acting either at a transcriptional or posttranscriptional level. The recent evidence that antisense repeat RNAs could modulate gene expression broadens our understanding of the pathogenic pathways and adds more complexity to the development of therapeutic strategies for these disorders. In this review, we cover the amazing progress made in the understanding of the pathogenic mechanisms associated with repeat expansion neurodegenerative and neuromuscular diseases with a focus on the impact of antisense repeat transcription in the development of efficient therapies.
Keywords: RNA foci; nuclear inclusions; splicing misregulation; trinucleotide repeats.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Zu T., Gibbens B., Doty N.S., Gomes-Pereira M., Huguet A., Stone M.D., Margolis J., Peterson M., Markowski T.W., Ingram M.A.C., et al. Non-ATG–initiated translation directed by microsatellite expansions. Proc. Natl. Acad. Sci. USA. 2010;108:260–265. doi: 10.1073/pnas.1013343108. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

