Closely Related Vibrio alginolyticus Strains Encode an Identical Repertoire of Caudovirales-Like Regions and Filamentous Phages
- PMID: 33261037
- PMCID: PMC7761403
- DOI: 10.3390/v12121359
Closely Related Vibrio alginolyticus Strains Encode an Identical Repertoire of Caudovirales-Like Regions and Filamentous Phages
Abstract
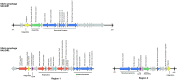
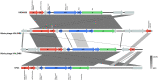
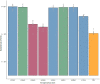
Many filamentous vibriophages encode virulence genes that lead to the emergence of pathogenic bacteria. Most genomes of filamentous vibriophages characterized up until today were isolated from human pathogens. Despite genome-based predictions that environmental Vibrios also contain filamentous phages that contribute to bacterial virulence, empirical evidence is scarce. This study aimed to characterize the bacteriophages of a marine pathogen, Vibrio alginolyticus (Kiel-alginolyticus ecotype) and to determine their role in bacterial virulence. To do so, we sequenced the phage-containing supernatant of eight different V. alginolyticus strains, characterized the phages therein and performed infection experiments on juvenile pipefish to assess their contribution to bacterial virulence. We were able to identify two actively replicating filamentous phages. Unique to this study was that all eight bacteria of the Kiel-alginolyticus ecotype have identical bacteriophages, supporting our previously established theory of a clonal expansion of the Kiel-alginolyticus ecotype. We further found that in one of the two filamentous phages, two phage-morphogenesis proteins (Zot and Ace) share high sequence similarity with putative toxins encoded on the Vibrio cholerae phage CTXΦ. The coverage of this filamentous phage correlated positively with virulence (measured in controlled infection experiments on the eukaryotic host), suggesting that this phage contributes to bacterial virulence.
Keywords: Inoviridae; Vibrio virulence; filamentous phages; prophages; vibriophages; zot.
Conflict of interest statement
The authors declare there are no conflict of interest.
Figures



References
-
- Chang B., Miyamoto H., Taniguchi H., Yoshida S.-I. Isolation and Genetic Characterization of a Novel Filamentous Bacteriophage, a Deleted Form of Phage f237, from a Pandemic Vibrio parahaemolyticus O4:K68 Strain. Microbiol. Immunol. 2002;46:565–569. doi: 10.1111/j.1348-0421.2002.tb02734.x. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

