Slow Adaptive Response of Budding Yeast Cells to Stable Conditions of Continuous Culture Can Occur without Genome Modifications
- PMID: 33261040
- PMCID: PMC7759791
- DOI: 10.3390/genes11121419
Slow Adaptive Response of Budding Yeast Cells to Stable Conditions of Continuous Culture Can Occur without Genome Modifications
Abstract
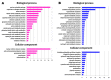
Continuous cultures assure the invariability of environmental conditions and the metabolic state of cultured microorganisms, whereas batch-cultured cells undergo constant changes in nutrients availability. For that reason, continuous culture is sometimes employed in the whole transcriptome, whole proteome, or whole metabolome studies. However, the typical method for establishing uniform growth of a cell population, i.e., by limited chemostat, results in the enrichment of the cell population gene pool with mutations adaptive for starvation conditions. These adaptive changes can skew the results of large-scale studies. It is commonly assumed that these adaptations reflect changes in the genome, and this assumption has been confirmed experimentally in rare cases. Here we show that in a population of budding yeast cells grown for over 200 generations in continuous culture in non-limiting minimal medium and therefore not subject to selection pressure, remodeling of transcriptome occurs, but not as a result of the accumulation of adaptive mutations. The observed changes indicate a shift in the metabolic balance towards catabolism, a decrease in ribosome biogenesis, a decrease in general stress alertness, reorganization of the cell wall, and transactions occurring at the cell periphery. These adaptive changes signify the acquisition of a new lifestyle in a stable nonstressful environment. The absence of underlying adaptive mutations suggests these changes may be regulated by another mechanism.
Keywords: Saccharomyces cerevisiae; adaptation; epigenetic; stress alertness; transcription factor; transcriptome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

