RNA sequencing analysis of altered expression of long noncoding RNAs associated with Schistosoma japonicum infection in the murine liver and spleen
- PMID: 33261628
- PMCID: PMC7705434
- DOI: 10.1186/s13071-020-04457-9
RNA sequencing analysis of altered expression of long noncoding RNAs associated with Schistosoma japonicum infection in the murine liver and spleen
Abstract
Background: Schistosomiasis is a chronic, debilitating infectious disease caused by members of the genus Schistosoma. Previous findings have suggested a relationship between infection with Schistosoma spp. and alterations in the liver and spleen of infected animals. Recent reports have shown the regulatory role of noncoding RNAs, such as long noncoding RNAs (lncRNAs), in different biological processes. However, little is known about the role of lncRNAs in the mouse liver and spleen during Schistosoma japonicum infection.
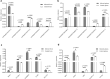
Methods: In this study, we identified and investigated lncRNAs using standard RNA sequencing (RNA-Seq). The biological functions of the altered expression of lncRNAs and their target genes were predicted using bioinformatics. Ten dysregulated lncRNAs were selected randomly and validated in reverse transcription-quantitative real-time polymerase chain reaction (RT-qPCR) experiments.
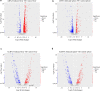
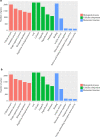
Results: Our study identified 29,845 and 33,788 lncRNAs from the liver and spleen, respectively, of which 212 were novel lncRNAs. We observed that 759 and 789 of the lncRNAs were differentially expressed in the respective organs. The RT-qPCR results correlated well with the sequencing data. In the liver, 657 differentially expressed lncRNAs were predicted to target 2548 protein-coding genes, whereas in the spleen 660 differentially expressed lncRNAs were predicted to target 2673 protein-coding genes. Moreover, functional annotation showed that the target genes of the differentially expressed lncRNAs were associated with cellular processes, metabolic processes, and binding, and were significantly enriched in metabolic pathways, the cell cycle, ubiquitin-mediated proteolysis, and pathways in cancer.
Conclusions: Our study showed that numerous lncRNAs were differentially expressed in S. japonicum-infected liver and spleen compared to control liver and spleen; this suggested that lncRNAs may be involved in pathogenesis in the liver and spleen during S. japonicum infection.
Keywords: Liver; Long noncoding RNA; Pathogenesis; RNA sequencing; Schistosoma japonicum; Spleen.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




Similar articles
-
[Screening and functional analysis of differentially expressed long non-coding RNA in the liver of mice infected with Schistosoma japonicum during the chronic pathogenic stage].Zhongguo Xue Xi Chong Bing Fang Zhi Za Zhi. 2024 May 15;36(2):137-147. doi: 10.16250/j.32.1374.2024076. Zhongguo Xue Xi Chong Bing Fang Zhi Za Zhi. 2024. PMID: 38857956 Chinese.
-
Schistosoma japonicum infection-mediated downregulation of lncRNA Malat1 contributes to schistosomiasis hepatic fibrosis by the Malat1/miR-96/Smad7 pathway.Parasit Vectors. 2024 Oct 3;17(1):413. doi: 10.1186/s13071-024-06499-9. Parasit Vectors. 2024. PMID: 39363237 Free PMC article.
-
[Whole transcriptome analysis and critical gene regulatory network analysis during Schistosoma japonicum infection and praziquantel treatment in mice].Zhongguo Xue Xi Chong Bing Fang Zhi Za Zhi. 2022 Apr 11;34(2):128-140. doi: 10.16250/j.32.1374.2021299. Zhongguo Xue Xi Chong Bing Fang Zhi Za Zhi. 2022. PMID: 35537834 Chinese.
-
Schistosoma japonicum infection in the pig as a model for human schistosomiasis japonica.Acta Trop. 2000 Sep 18;76(2):85-99. doi: 10.1016/s0001-706x(00)00103-0. Acta Trop. 2000. PMID: 10936567 Review.
-
Can Schistosoma japonicum infection cause liver cancer?J Helminthol. 2025 Feb 10;99:e11. doi: 10.1017/S0022149X24000762. J Helminthol. 2025. PMID: 39924660 Review.
Cited by
-
Serum proteomic profiling in patients with advanced Schistosoma japonicum-induced hepatic fibrosis.Parasit Vectors. 2021 May 1;14(1):232. doi: 10.1186/s13071-021-04734-1. Parasit Vectors. 2021. PMID: 33933138 Free PMC article.
-
Ikzf2 Regulates the Development of ICOS+ Th Cells to Mediate Immune Response in the Spleen of S. japonicum-Infected C57BL/6 Mice.Front Immunol. 2021 Aug 12;12:687919. doi: 10.3389/fimmu.2021.687919. eCollection 2021. Front Immunol. 2021. PMID: 34475870 Free PMC article.
-
Both host and parasite non-coding RNAs co-ordinate the regulation of macrophage gene expression to reduce pro-inflammatory immune responses and promote tissue repair pathways during infection with fasciola hepatica.RNA Biol. 2024 Jan;21(1):62-77. doi: 10.1080/15476286.2024.2408706. Epub 2024 Sep 30. RNA Biol. 2024. PMID: 39344634 Free PMC article.
-
Knocking Down Gm16685 Decreases Liver Granuloma in Murine Schistosomiasis Japonica.Microorganisms. 2023 Mar 21;11(3):796. doi: 10.3390/microorganisms11030796. Microorganisms. 2023. PMID: 36985369 Free PMC article.
-
Long Non-Coding RNA and mRNA Expression Analysis in Liver of Mice With Clonorchis sinensis Infection.Front Cell Infect Microbiol. 2022 Jan 19;11:754224. doi: 10.3389/fcimb.2021.754224. eCollection 2021. Front Cell Infect Microbiol. 2022. PMID: 35127549 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

