Production of Noncapped Genomic RNAs Is Critical to Sindbis Virus Disease and Pathogenicity
- PMID: 33262258
- PMCID: PMC7733944
- DOI: 10.1128/mBio.02675-20
Production of Noncapped Genomic RNAs Is Critical to Sindbis Virus Disease and Pathogenicity
Abstract
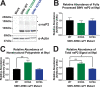
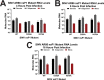
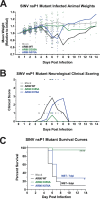
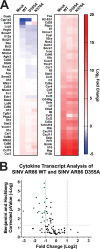
Alphaviruses are positive-sense RNA viruses that utilize a 5' cap structure to facilitate translation of viral proteins and to protect the viral RNA genome. Nonetheless, significant quantities of viral genomic RNAs that lack a canonical 5' cap structure are produced during alphaviral replication and packaged into viral particles. However, the role/impact of the noncapped genomic RNA (ncgRNA) during alphaviral infection in vivo has yet to be characterized. To determine the importance of the ncgRNA in vivo, the previously described D355A and N376A nsP1 mutations, which increase or decrease nsP1 capping activity, respectively, were incorporated into the neurovirulent AR86 strain of Sindbis virus to enable characterization of the impact of altered capping efficiency in a murine model of infection. Mice infected with the N376A nsP1 mutant exhibited slightly decreased rates of mortality and delayed weight loss and neurological symptoms, although levels of inflammation in the brain were similar to those of wild-type infection. Although the D355A mutation resulted in decreased antiviral gene expression and increased resistance to interferon in vitro, mice infected with the D355A mutant showed significantly reduced mortality and morbidity compared to mice infected with wild-type virus. Interestingly, expression of proinflammatory cytokines was found to be significantly decreased in mice infected with the D355A mutant, suggesting that capping efficiency and the production of ncgRNA are vital to eliciting pathogenic levels of inflammation. Collectively, these data indicate that the ncgRNA have important roles during alphaviral infection and suggest a novel mechanism by which noncapped viral RNAs aid in viral pathogenesis.IMPORTANCE Mosquito-transmitted alphaviruses have been the cause of widespread outbreaks of disease that can range from mild illness to lethal encephalitis or severe polyarthritis. There are currently no safe and effective vaccines or therapeutics with which to prevent or treat alphaviral disease, highlighting the need to better understand alphaviral pathogenesis to develop novel antiviral strategies. This report reveals production of noncapped genomic RNAs (ncgRNAs) to be a novel determinant of alphaviral virulence and offers insight into the importance of inflammation to pathogenesis. Taken together, the findings reported here suggest that the ncgRNAs contribute to alphaviral pathogenesis through the sensing of the ncgRNAs during alphaviral infection and are necessary for the development of severe disease.
Keywords: RNA virus; alphavirus; capping; nsP1; viral pathogenesis.
Copyright © 2020 LaPointe et al.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

