Plasmid-Mediated AmpC β-Lactamase CITM and DHAM Genes Among Gram-Negative Clinical Isolates
- PMID: 33262619
- PMCID: PMC7699442
- DOI: 10.2147/IDR.S284751
Plasmid-Mediated AmpC β-Lactamase CITM and DHAM Genes Among Gram-Negative Clinical Isolates
Abstract
Background: Antibiotic resistance mediated by the production of extended-spectrum β-lactamases (ESBLs) and AmpC β-lactamases is posing a serious threat in the management of the infections caused by Gram-negative pathogens. The aim of this study was to determine the prevalence of two AmpC β-lactamases genes, bla CITM and bla DHAM, in Gram-negative bacterial isolates.
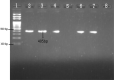
Materials and methods: A total of 1151 clinical samples were obtained and processed at the microbiology laboratory of Annapurna Neurological Institute and Allied Science, Kathmandu between June 2017 and January 2018. Gram-negative isolates thus obtained were tested for antimicrobial susceptibility testing (AST) using Kirby-Bauer disk diffusion method. AmpC β-lactamase production was detected by disk approximation method using phenylboronic acid (PBA). Confirmed AmpC β-lactamase producers were further screened for bla CITM and bla DHAM genes by conventional polymerase chain reaction (PCR).
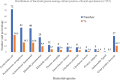
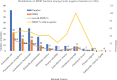
Results: Out of 1151 clinical specimens, 22% (253/1152) had bacterial growth. Of the total isolates, 89.3% (226/253) were Gram-negatives, with E. coli as the most predominant species (n=72) followed by Pseudomonas aeruginosa (n=41). In the AST, 46.9% (106/226) of the Gram-negative isolates were multidrug resistant (MDR). In disk diffusion test, 113 (50%) isolates showed resistance against cefoxitin, among which 91 isolates (83 by disk test and Boronic acid test, 8 by Boronic test only) were confirmed as AmpC β-lactamase-producers. In PCR assay, 90.1% (82/91) and 87.9% (80/91) of the isolates tested positive for production of bla CITM and bla DHAM genes, respectively.
Conclusions: High prevalence of AmpC β-lactamase-producers in our study is an alarming sign. This study recommends the use of modern diagnostic facilities in the clinical settings for early detection and management which can optimize the treatment therapies, curb the growth and spread of the drug-resistant pathogens.
Keywords: AmpC β-lactamase; ESBLs; MDR; blaCITM; blaDHAM; polymerase chain reaction.
© 2020 Aryal et al.
Conflict of interest statement
All the authors hereby declare that they have no competing interests.
Figures

References
LinkOut - more resources
Full Text Sources

