Unraveling Pseudomonas aeruginosa and Candida albicans Communication in Coinfection Scenarios: Insights Through Network Analysis
- PMID: 33262953
- PMCID: PMC7686562
- DOI: 10.3389/fcimb.2020.550505
Unraveling Pseudomonas aeruginosa and Candida albicans Communication in Coinfection Scenarios: Insights Through Network Analysis
Abstract
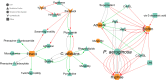
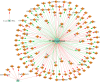
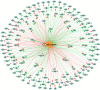
Modern medicine is currently facing huge setbacks concerning infection therapeutics as microorganisms are consistently knocking down every antimicrobial wall set before them. The situation becomes more worrying when taking into account that, in both environmental and disease scenarios, microorganisms present themselves as biofilm communities that are often polymicrobial. This comprises a competitive advantage, with interactions between different species altering host responses, antimicrobial effectiveness, microbial pathogenesis and virulence, usually augmenting the severity of the infection and contributing for the recalcitrance towards conventional therapy. Pseudomonas aeruginosa and Candida albicans are two opportunistic pathogens often co-isolated from infections, mainly from mucosal tissues like the lung. Despite the billions of years of co-existence, this pair of microorganisms is a great example on how little is known about cross-kingdom interactions, particularly within the context of coinfections. Given the described scenario, this study aimed to collect, curate, and analyze all published experimental information on the molecular basis of P. aeruginosa and C. albicans interactions in biofilms, in order to shed light into key mechanisms that may affect infection prognosis, increasing this area of knowledge. Publications were optimally retrieved from PubMed and Web of Science and classified as to their relevance. Data was then systematically and manually curated, analyzed, and further reconstructed as networks. A total of 641 interactions between the two pathogens were annotated, outputting knowledge on important molecular players affecting key virulence mechanisms, such as hyphal growth, and related genes and proteins, constituting potential therapeutic targets for infections related to these bacterial-fungal consortia. Contrasting interactions were also analyzed, and quorum-sensing inhibition approaches were highlighted. All annotated data was made publicly available at www.ceb.uminho.pt/ISCTD, a database already containing similar data for P. aeruginosa and Staphylococcus aureus communication. This will allow researchers to cut on time and effort when studying this particular subject, facilitating the understanding of the basis of the inter-species and inter-kingdom interactions and how it can be modulated to help design alternative and more effective tailored therapies. Finally, data deposition will serve as base for future dataset integration, whose analysis will hopefully give insights into communications in more complex and varied biofilm communities.
Keywords: Candida albicans; Pseudomonas aeruginosa; biofilms; coinfection; database; interactions; polymicrobial.
Copyright © 2020 Grainha, Jorge, Alves, Lopes and Pereira.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

