Genomic Prediction of Additive and Dominant Effects on Wool and Blood Traits in Alpine Merino Sheep
- PMID: 33263012
- PMCID: PMC7686030
- DOI: 10.3389/fvets.2020.573692
Genomic Prediction of Additive and Dominant Effects on Wool and Blood Traits in Alpine Merino Sheep
Abstract
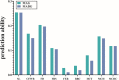
Dominant genetic effects may provide a critical contribution to the total genetic variation of quantitative and complex traits. However, investigations of genome-wide markers to study the genomic prediction (GP) and genetic mechanisms of complex traits generally ignore dominant genetic effects. The increasing availability of genomic datasets and the potential benefits of the inclusion of non-additive genetic effects in GP have recently renewed attention to incorporation of these effects in genomic prediction models. In the present study, data from 498 genotyped Alpine Merino sheep were adopted to estimate the additive and dominant genetic effects of 9 wool and blood traits via two linear models: (1) an additive effect model (MAG) and (2) a model that included both additive and dominant genetic effects (MADG). Moreover, a method of 5-fold cross validation was used to evaluate the capability of GP in the two different models. The results of variance component estimates for each trait suggested that for fleece extension rate (73%), red blood cell count (28%), and hematocrit (25%), a large component of phenotypic variation was explained by dominant genetic effects. The results of cross validation demonstrated that the MADG model, comprising additive and dominant genetic effects, did not display an apparent advantage over the MAG model that included only additive genetic effects, i.e., the model that included dominant genetic effects did not improve the capability for prediction of the genomic model. Consequently, inclusion of dominant effects in the GP model may not be beneficial for wool and blood traits in the population of Alpine Merino sheep.
Keywords: Alpine Merino sheep; additive effects; dominant effects; genomic prediction; prediction accuracy.
Copyright © 2020 Zhu, Zhao, Han, Yuan, Guo, Liu, Yue, Qiao, Wang, Li, Gun and Yang.
Figures
Similar articles
-
Estimates of (co)variance components and phenotypic and genetic parameters of growth traits and wool traits in Alpine Merino sheep.J Anim Breed Genet. 2022 May;139(3):351-365. doi: 10.1111/jbg.12669. Epub 2022 Feb 16. J Anim Breed Genet. 2022. PMID: 35170817
-
Evaluation of Bayesian alphabet and GBLUP based on different marker density for genomic prediction in Alpine Merino sheep.G3 (Bethesda). 2021 Oct 19;11(11):jkab206. doi: 10.1093/g3journal/jkab206. G3 (Bethesda). 2021. PMID: 34849779 Free PMC article.
-
Genomic Selection for Live Weight in the 14th Month in Alpine Merino Sheep Combining GWAS Information.Animals (Basel). 2023 Nov 14;13(22):3516. doi: 10.3390/ani13223516. Animals (Basel). 2023. PMID: 38003134 Free PMC article.
-
Multiple-trait QTL mapping and genomic prediction for wool traits in sheep.Genet Sel Evol. 2017 Aug 15;49(1):62. doi: 10.1186/s12711-017-0337-y. Genet Sel Evol. 2017. PMID: 28810834 Free PMC article.
-
The Effect of Integrating Genomic Information into Genetic Evaluations of Chinese Merino Sheep.Animals (Basel). 2020 Mar 28;10(4):569. doi: 10.3390/ani10040569. Animals (Basel). 2020. PMID: 32231053 Free PMC article.
Cited by
-
Improvement in genetic evaluation of quantitative traits in sheep by enriching genetic model with dominance effects.Sci Rep. 2025 Aug 17;15(1):30144. doi: 10.1038/s41598-025-16005-5. Sci Rep. 2025. PMID: 40820152 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials