The Legacy of Recurrent Introgression during the Radiation of Hares
- PMID: 33263746
- PMCID: PMC8048390
- DOI: 10.1093/sysbio/syaa088
The Legacy of Recurrent Introgression during the Radiation of Hares
Abstract
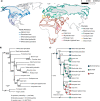
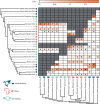
Hybridization may often be an important source of adaptive variation, but the extent and long-term impacts of introgression have seldom been evaluated in the phylogenetic context of a radiation. Hares (Lepus) represent a widespread mammalian radiation of 32 extant species characterized by striking ecological adaptations and recurrent admixture. To understand the relevance of introgressive hybridization during the diversification of Lepus, we analyzed whole exome sequences (61.7 Mb) from 15 species of hares (1-4 individuals per species), spanning the global distribution of the genus, and two outgroups. We used a coalescent framework to infer species relationships and divergence times, despite extensive genealogical discordance. We found high levels of allele sharing among species and show that this reflects extensive incomplete lineage sorting and temporally layered hybridization. Our results revealed recurrent introgression at all stages along the Lepus radiation, including recent gene flow between extant species since the last glacial maximum but also pervasive ancient introgression occurring since near the origin of the hare lineages. We show that ancient hybridization between northern hemisphere species has resulted in shared variation of potential adaptive relevance to highly seasonal environments, including genes involved in circadian rhythm regulation, pigmentation, and thermoregulation. Our results illustrate how the genetic legacy of ancestral hybridization may persist across a radiation, leaving a long-lasting signature of shared genetic variation that may contribute to adaptation. [Adaptation; ancient introgression; hybridization; Lepus; phylogenomics.].
© The Author(s) 2020. Published by Oxford University Press, on behalf of the Society of Systematic Biologists.
Figures



References
-
- Alves P.C., Melo-Ferreira J., Branco M., Suchentrunk F., Ferrand N., Harris D.J.. 2008. Evidence for genetic similarity of two allopatric European hares (Lepus corsicanus and L. castroviejoi) inferred from nuclear DNA sequences. Mol. Phylogenet. Evol. 46:1191–1197. - PubMed
-
- Barlow A., Cahill J.A., Hartmann S., Theunert C., Xenikoudakis G., Fortes G.G., Paijmans J.L.A., Rabeder G., Frischauf C., Grandal-d’Anglade A., García-Vázquez A., Murtskhvaladze M., Saarma U., Anijalg P., Skrbinšek T., Bertorelle G., Gasparian B., Bar-Oz G., Pinhasi R., Slatkin M., Dalén L., Shapiro B., Hofreiter M.. 2018. Partial genomic survival of cave bears in living brown bears. Nat. Ecol. Evol. 2:1563–1570. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

