Ion Binding Properties and Dynamics of the bcl- 2 G-Quadruplex Using a Polarizable Force Field
- PMID: 33264004
- PMCID: PMC7775346
- DOI: 10.1021/acs.jcim.0c01064
Ion Binding Properties and Dynamics of the bcl- 2 G-Quadruplex Using a Polarizable Force Field
Abstract
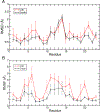
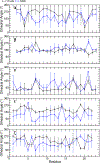
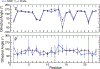
G-quadruplexes (GQs) are topologically diverse, highly thermostable noncanonical nucleic acid structures that form in guanine-rich sequences in DNA and RNA. GQs are implicated in transcriptional and translational regulation and genome maintenance, and deleterious alterations to their structures contribute to diseases such as cancer. The expression of the B-cell lymphoma 2 (Bcl-2) antiapoptotic protein, for example, is under transcriptional control of a GQ in the promoter of the bcl-2 gene. Modulation of the bcl-2 GQ by small molecules is of interest for chemotherapeutic development but doing so requires knowledge of the factors driving GQ folding and stabilization. To develop a greater understanding of the electrostatic properties of the bcl-2 promoter GQ, we performed molecular dynamics simulations using the Drude-2017 polarizable force field and compared relevant outcomes to the nonpolarizable CHARMM36 force field. Our simulation outcomes highlight the importance of dipole-dipole interactions in the bcl-2 GQ, particularly during the recruitment of a bulk K+ ion to the solvent-exposed face of the tetrad stem. We also predict and characterize an "electronegative pocket" at the tetrad-long loop junction that induces local backbone conformational change and may induce local conformational changes at cellular concentrations of K+. These outcomes suggest that moieties within the bcl-2 GQ can be targeted by small molecules to modulate bcl-2 GQ stability.
Figures
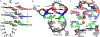




Similar articles
-
Same fold, different properties: polarizable molecular dynamics simulations of telomeric and TERRA G-quadruplexes.Nucleic Acids Res. 2020 Jan 24;48(2):561-575. doi: 10.1093/nar/gkz1154. Nucleic Acids Res. 2020. PMID: 31807754 Free PMC article.
-
Polarizable Molecular Dynamics Simulations of Two c-kit Oncogene Promoter G-Quadruplexes: Effect of Primary and Secondary Structure on Loop and Ion Sampling.J Chem Theory Comput. 2020 May 12;16(5):3430-3444. doi: 10.1021/acs.jctc.0c00191. Epub 2020 Apr 30. J Chem Theory Comput. 2020. PMID: 32307997 Free PMC article.
-
Cation competition and recruitment around the c-kit1 G-quadruplex using polarizable simulations.Biophys J. 2021 Jun 1;120(11):2249-2261. doi: 10.1016/j.bpj.2021.03.022. Epub 2021 Mar 29. Biophys J. 2021. PMID: 33794153 Free PMC article.
-
Folding of guanine quadruplex molecules-funnel-like mechanism or kinetic partitioning? An overview from MD simulation studies.Biochim Biophys Acta Gen Subj. 2017 May;1861(5 Pt B):1246-1263. doi: 10.1016/j.bbagen.2016.12.008. Epub 2016 Dec 13. Biochim Biophys Acta Gen Subj. 2017. PMID: 27979677 Review.
-
G-quadruplex dynamics.Biochim Biophys Acta Proteins Proteom. 2017 Nov;1865(11 Pt B):1544-1554. doi: 10.1016/j.bbapap.2017.06.012. Epub 2017 Jun 20. Biochim Biophys Acta Proteins Proteom. 2017. PMID: 28642152 Review.
Cited by
-
Differences in Conformational Sampling and Intrinsic Electric Fields Drive Ion Binding in Telomeric and TERRA G-Quadruplexes.J Chem Inf Model. 2023 Nov 13;63(21):6851-6862. doi: 10.1021/acs.jcim.3c01305. Epub 2023 Oct 17. J Chem Inf Model. 2023. PMID: 37847037 Free PMC article.
-
Combining Electrospray Mass Spectrometry (ESI-MS) and Computational Techniques in the Assessment of G-Quadruplex Ligands: A Hybrid Approach to Optimize Hit Discovery.J Med Chem. 2021 Sep 23;64(18):13174-13190. doi: 10.1021/acs.jmedchem.1c00962. Epub 2021 Sep 12. J Med Chem. 2021. PMID: 34510895 Free PMC article.
-
Base pair dynamics, electrostatics, and thermodynamics at the LTR-III quadruplex:duplex junction.Biophys J. 2024 May 7;123(9):1129-1138. doi: 10.1016/j.bpj.2024.03.042. Epub 2024 Apr 4. Biophys J. 2024. PMID: 38576161 Free PMC article.
-
Ion-Dependent Conformational Plasticity of Telomeric G-Hairpins and G-Quadruplexes.ACS Omega. 2022 Jun 29;7(27):23368-23379. doi: 10.1021/acsomega.2c01600. eCollection 2022 Jul 12. ACS Omega. 2022. PMID: 35847338 Free PMC article.
-
Simulations predict preferred Mg2+ coordination in a nonenzymatic primer-extension reaction center.Biophys J. 2024 Jun 18;123(12):1579-1591. doi: 10.1016/j.bpj.2024.04.032. Epub 2024 May 3. Biophys J. 2024. PMID: 38702884 Free PMC article.
References
-
- Phan AT; Kuryavyi V; Darnell JC; Serganov A; Majumdar A; Ilin S; Raslin T; Polonskaia A; Chen C; Clain D; Darnell RB; Patel DJ Structure-Function Studies of FMRP RGG Peptide Recognition of an RNA Duplex-Quadruplex Junction. Nat. Struct. Mol. Biol. 2011, 18 (7), 796–804. 10.1038/nsmb.2064. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

