The structural basis of accelerated host cell entry by SARS-CoV-2†
- PMID: 33264497
- PMCID: PMC7753708
- DOI: 10.1111/febs.15651
The structural basis of accelerated host cell entry by SARS-CoV-2†
Abstract
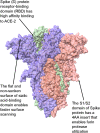
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the causative agent of the pandemic coronavirus disease 2019 (COVID-19) that exhibits an overwhelming contagious capacity over other human coronaviruses (HCoVs). This structural snapshot describes the structural bases underlying the pandemic capacity of SARS-CoV-2 and explains its fast motion over respiratory epithelia that allow its rapid cellular entry. Based on notable viral spike (S) protein features, we propose that the flat sialic acid-binding domain at the N-terminal domain (NTD) of the S1 subunit leads to more effective first contact and interaction with the sialic acid layer over the epithelium, and this, in turn, allows faster viral 'surfing' of the epithelium and receptor scanning by SARS-CoV-2. Angiotensin-converting enzyme 2 (ACE-2) protein on the epithelial surface is the primary entry receptor for SARS-CoV-2, and protein-protein interaction assays demonstrate high-affinity binding of the spike protein (S protein) to ACE-2. To date, no high-frequency mutations were detected at the C-terminal domain of the S1 subunit in the S protein, where the receptor-binding domain (RBD) is located. Tight binding to ACE-2 by a conserved viral RBD suggests the ACE2-RBD interaction is likely optimal. Moreover, the viral S subunit contains a cleavage site for furin and other proteases, which accelerates cell entry by SARS-CoV-2. The model proposed here describes a structural basis for the accelerated host cell entry by SARS-CoV-2 relative to other HCoVs and also discusses emerging hypotheses that are likely to contribute to the development of antiviral strategies to combat the pandemic capacity of SARS-CoV-2.
Keywords: COVID-19; SARS-CoV-2; furin protease; receptor-binding domain; sialic acid-binding domain.
© 2020 Federation of European Biochemical Societies.
Conflict of interest statement
MS has a utility model application with the file number of GM 63/2020 to the Österreichisches Patentamt.
Figures


References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

